IFNα induces CCR5 in CD4+ T cells of HIV patients causing pathogenic elevation
- PMID: 38504093
- PMCID: PMC10951336
- DOI: 10.1038/s43856-024-00453-7
IFNα induces CCR5 in CD4+ T cells of HIV patients causing pathogenic elevation
Abstract
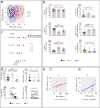
Background: Among people living with HIV, elite controllers (ECs) maintain an undetectable viral load, even without receiving anti-HIV therapy. In non-EC patients, this therapy leads to marked improvement, including in immune parameters, but unlike ECs, non-EC patients still require ongoing treatment and experience co-morbidities. In-depth, comprehensive immune analyses comparing EC and treated non-EC patients may reveal subtle, consistent differences. This comparison could clarify whether elevated circulating interferon-alpha (IFNα) promotes widespread immune cell alterations and persists post-therapy, furthering understanding of why non-EC patients continue to need treatment.
Methods: Levels of IFNα in HIV-infected EC and treated non-EC patients were compared, along with blood immune cell subset distribution and phenotype, and functional capacities in some cases. In addition, we assessed mechanisms potentially associated with IFNα overload.
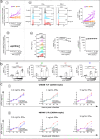
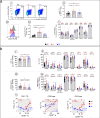
Results: Treatment of non-EC patients results in restoration of IFNα control, followed by marked improvement in distribution numbers, phenotypic profiles of blood immune cells, and functional capacity. These changes still do not lead to EC status, however, and IFNα can induce these changes in normal immune cell counterparts in vitro. Hypothesizing that persistent alterations could arise from inalterable effects of IFNα at infection onset, we verified an IFNα-related mechanism. The protein induces the HIV coreceptor CCR5, boosting HIV infection and reducing the effects of anti-HIV therapies. EC patients may avoid elevated IFNα following on infection with a lower inoculum of HIV or because of some unidentified genetic factor.
Conclusions: Early control of IFNα is essential for better prognosis of HIV-infected patients.
Plain language summary
The treatment for HIV, known as antiretroviral therapy (ART), does not cure HIV but enables individuals to live longer, healthier lives. In this study, we compared immune responses between elite controllers (ECs), who control their HIV infection without any treatment, and ART-treated and untreated patients. We demonstrate that IFNα, a small protein crucial in controlling immune system, is excessively produced at the onset of HIV infection and at levels that persist, resulting in poor HIV control without therapy. We show a mechanism for lack of control of HIV by IFNα. While inhibiting HIV, IFNα also simultaneously increases the HIV co-receptor, CCR5, thereby facilitating virus entry into the target cell. This is avoided by ECs which we hypothesize is associated with a lower infectious inoculum of HIV.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
IFNα induces CCR5 in CD4+ T-cells, causing its anti- HIV inefficiency and its subsequent pathogenic elevation, partially controlled by anti-HIV therapy.Res Sq [Preprint]. 2023 May 11:rs.3.rs-2813616. doi: 10.21203/rs.3.rs-2813616/v1. Res Sq. 2023. Update in: Commun Med (Lond). 2024 Mar 19;4(1):52. doi: 10.1038/s43856-024-00453-7. PMID: 37214795 Free PMC article. Updated. Preprint.
References
-
- Zhou, Q. et al. Malignancies in people living with HIV. AIDS Rev. 10.24875/AIDSRev.21000057 (2022). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

