Insulin signalling regulates Pink1 mRNA localization via modulation of AMPK activity to support PINK1 function in neurons
- PMID: 38504131
- PMCID: PMC10963278
- DOI: 10.1038/s42255-024-01007-w
Insulin signalling regulates Pink1 mRNA localization via modulation of AMPK activity to support PINK1 function in neurons
Abstract
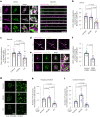
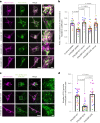
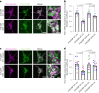
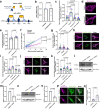
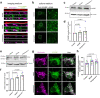
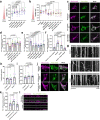
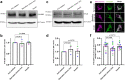
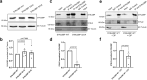
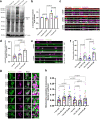
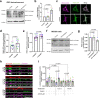
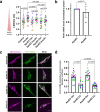
Mitochondrial quality control failure is frequently observed in neurodegenerative diseases. The detection of damaged mitochondria by stabilization of PTEN-induced kinase 1 (PINK1) requires transport of Pink1 messenger RNA (mRNA) by tethering it to the mitochondrial surface. Here, we report that inhibition of AMP-activated protein kinase (AMPK) by activation of the insulin signalling cascade prevents Pink1 mRNA binding to mitochondria. Mechanistically, AMPK phosphorylates the RNA anchor complex subunit SYNJ2BP within its PDZ domain, a phosphorylation site that is necessary for its interaction with the RNA-binding protein SYNJ2. Notably, loss of mitochondrial Pink1 mRNA association upon insulin addition is required for PINK1 protein activation and its function as a ubiquitin kinase in the mitophagy pathway, thus placing PINK1 function under metabolic control. Induction of insulin resistance in vitro by the key genetic Alzheimer risk factor apolipoprotein E4 retains Pink1 mRNA at the mitochondria and prevents proper PINK1 activity, especially in neurites. Our results thus identify a metabolic switch controlling Pink1 mRNA localization and PINK1 activity via insulin and AMPK signalling in neurons and propose a mechanistic connection between insulin resistance and mitochondrial dysfunction.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Valente EM, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. - PubMed
MeSH terms
Substances
Grants and funding
- MPRGL/Max-Planck-Gesellschaft (Max Planck Society)
- EXC 2145 SyNergy - ID 390857198 and HA 7728/2-1 - ID 453679203/Deutsche Forschungsgemeinschaft (German Research Foundation)
- ERC StG 463 Project 101077138 - MitoPIP/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Research Materials

