Salt tolerance evaluation and mini-core collection development in Miscanthus sacchariflorus and M. lutarioriparius
- PMID: 38504893
- PMCID: PMC10948507
- DOI: 10.3389/fpls.2024.1364826
Salt tolerance evaluation and mini-core collection development in Miscanthus sacchariflorus and M. lutarioriparius
Abstract
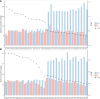
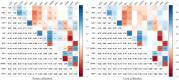
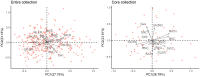
Marginal lands, such as those with saline soils, have potential as alternative resources for cultivating dedicated biomass crops used in the production of renewable energy and chemicals. Optimum utilization of marginal lands can not only alleviate the competition for arable land use with primary food crops, but also contribute to bioenergy products and soil improvement. Miscanthus sacchariflorus and M. lutarioriparius are prominent perennial plants suitable for sustainable bioenergy production in saline soils. However, their responses to salt stress remain largely unexplored. In this study, we utilized 318 genotypes of M. sacchariflorus and M. lutarioriparius to assess their salt tolerance levels under 150 mM NaCl using 14 traits, and subsequently established a mini-core elite collection for salt tolerance. Our results revealed substantial variation in salt tolerance among the evaluated genotypes. Salt-tolerant genotypes exhibited significantly lower Na+ content, and K+ content was positively correlated with Na+ content. Interestingly, a few genotypes with higher Na+ levels in shoots showed improved shoot growth characteristics. This observation suggests that M. sacchariflorus and M. lutarioriparius adapt to salt stress by regulating ion homeostasis, primarily through enhanced K+ uptake, shoot Na+ exclusion, and Na+ sequestration in shoot vacuoles. To evaluate salt tolerance comprehensively, we developed an assessment value (D value) based on the membership function values of the 14 traits. We identified three highly salt-tolerant, 50 salt-tolerant, 127 moderately salt-tolerant, 117 salt-sensitive, and 21 highly salt-sensitive genotypes at the seedling stage by employing the D value. A mathematical evaluation model for salt tolerance was established for M. sacchariflorus and M. lutarioriparius at the seedling stage. Notably, the mini-core collection containing 64 genotypes developed using the Core Hunter algorithm effectively represented the overall variability of the entire collection. This mini-core collection serves as a valuable gene pool for future in-depth investigations of salt tolerance mechanisms in Miscanthus.
Keywords: Miscanthus lutarioriparius; Miscanthus sacchariflorus; comprehensive evaluation; core collection; ion homeostasis; salt tolerance; seedling stage.
Copyright © 2024 Tang, Li, Zerpa-Catanho, Zhang, Yang, Zheng, Xue, Kuang, Liu, He, Yi and Xiao.
Conflict of interest statement
Author XH is employed by Hunan Heyi Crop Science Co., Ltd., Changsha, Hunan, China. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Genetic Diversity of Salt Tolerance in Miscanthus.Front Plant Sci. 2017 Feb 14;8:187. doi: 10.3389/fpls.2017.00187. eCollection 2017. Front Plant Sci. 2017. PMID: 28261243 Free PMC article.
-
Development of energy plants from hybrids between Miscanthus sacchariflorus and M. lutarioriparius grown on reclaimed mine land in the Loess Plateau of China.Front Plant Sci. 2023 Jan 16;13:1017712. doi: 10.3389/fpls.2022.1017712. eCollection 2022. Front Plant Sci. 2023. PMID: 36726684 Free PMC article.
-
Population structure of Miscanthus sacchariflorus reveals two major polyploidization events, tetraploid-mediated unidirectional introgression from diploid M. sinensis, and diversity centred around the Yellow Sea.Ann Bot. 2019 Oct 29;124(4):731-748. doi: 10.1093/aob/mcy161. Ann Bot. 2019. PMID: 30247525 Free PMC article.
-
Transcriptomic Characterization of Miscanthus sacchariflorus × M. lutarioriparius and Its Implications for Energy Crop Development in the Semiarid Mine Area.Plants (Basel). 2022 Jun 14;11(12):1568. doi: 10.3390/plants11121568. Plants (Basel). 2022. PMID: 35736719 Free PMC article.
-
Review of Current Prospects for Using Miscanthus-Based Polymers.Polymers (Basel). 2023 Jul 20;15(14):3097. doi: 10.3390/polym15143097. Polymers (Basel). 2023. PMID: 37514486 Free PMC article. Review.
Cited by
-
Screening and identification of grain sorghum germplasm for salt tolerance at seedling stage.Front Plant Sci. 2025 Jun 20;16:1610685. doi: 10.3389/fpls.2025.1610685. eCollection 2025. Front Plant Sci. 2025. PMID: 40621050 Free PMC article.
-
Omics-Driven Strategies for Developing Saline-Smart Lentils: A Comprehensive Review.Int J Mol Sci. 2024 Oct 22;25(21):11360. doi: 10.3390/ijms252111360. Int J Mol Sci. 2024. PMID: 39518913 Free PMC article. Review.
-
Elucidating the molecular basis of salt tolerance in potatoes through miRNA expression and phenotypic analysis.Sci Rep. 2025 Jan 21;15(1):2635. doi: 10.1038/s41598-025-86276-5. Sci Rep. 2025. PMID: 39838055 Free PMC article.
References
-
- Aravind J., Kaur V., Wankhede D. P., Nanjundan J. (2022). EvaluateCore: Quality Evaluation of Core Collections (R package version 0.1.3; ). Available at: https://aravind-j.github.io/EvaluateCore/, https://CRAN.R-project.org/package=EvaluateCore.
-
- Ashraf M. Y., Awan A. R., Mahmood K. (2012). Rehabilitation of saline ecosystems through cultivation of salt tolerant plants. Pak. J. Bot. 44, 69–75.
LinkOut - more resources
Full Text Sources

