Comparative proteomic analysis of tail regeneration in the green anole lizard, Anolis carolinensis
- PMID: 38505006
- PMCID: PMC10947082
- DOI: 10.1002/ntls.20210421
Comparative proteomic analysis of tail regeneration in the green anole lizard, Anolis carolinensis
Abstract
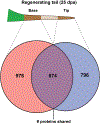
As amniote vertebrates, lizards are the most closely related organisms to humans capable of appendage regeneration. Lizards can autotomize, or release their tails as a means of predator evasion, and subsequently regenerate a functional replacement. Green anoles (Anolis carolinensis) can regenerate their tails through a process that involves differential expression of hundreds of genes, which has previously been analyzed by transcriptomic and microRNA analysis. To investigate protein expression in regenerating tissue, we performed whole proteomic analysis of regenerating tail tip and base. This is the first proteomic data set available for any anole lizard. We identified a total of 2,646 proteins - 976 proteins only in the regenerating tail base, 796 only in the tail tip, and 874 in both tip and base. For over 90% of these proteins in these tissues, we were able to assign a clear orthology to gene models in either the Ensembl or NCBI databases. For 13 proteins in the tail base, 9 proteins in the tail tip, and 10 proteins in both regions, the gene model in Ensembl and NCBI matched an uncharacterized protein, confirming that these predictions are present in the proteome. Ontology and pathways analysis of proteins expressed in the regenerating tail base identified categories including actin filament-based process, ncRNA metabolism, regulation of phosphatase activity, small GTPase mediated signal transduction, and cellular component organization or biogenesis. Analysis of proteins expressed in the tail tip identified categories including regulation of organelle organization, regulation of protein localization, ubiquitin-dependent protein catabolism, small GTPase mediated signal transduction, morphogenesis of epithelium, and regulation of biological quality. These proteomic findings confirm pathways and gene families activated in tail regeneration in the green anole as well as identify uncharacterized proteins whose role in regrowth remains to be revealed. This study demonstrates the insights that are possible from the integration of proteomic and transcriptomic data in tail regrowth in the green anole, with potentially broader application to studies in other regenerative models.
Keywords: Anole; Anolis carolinensis; Lizard; Proteome; Proteomic; Regeneration; Reptile; Squamate; Tail.
Figures






References
-
- Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, Ray DA, Boissinot S, Shedlock AM, Botka C, Castoe TA, Colbourne JK, Fujita MK, Moreno RG, Ten Hallers BF, Haussler D, Heger A, Heiman D, Janes DE, Johnson J, De Jong PJ, Koriabine MY, Lara M, Novick PA, Organ CL, Peach SE, Poe S, Pollock DD, De Queiroz K, Sanger T, Searle S, Smith JD, Smith Z, Swofford R, Turner-Maier J, Wade J, Young S, Zadissa A, Edwards SV., Glenn TC, Schneider CJ, Losos JB, Lander ES, Breen M, Ponting CP, Lindblad-Toh K. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477:587–591. DOI: 10.1038/nature10390. - DOI - PMC - PubMed
-
- Alibardi L 2018. Hyaluronate likely contributes to the immunesuppression of the regenerating tail blastema in lizards: Implications for organ regeneration in amniotes. Acta Zoologica. 99(4):321–330. DOI: 10.1111/azo.12214 - DOI
-
- Alibardi L 2017. Biological and molecular differences between tail regeneration and limb scarring in lizard: an inspiring model addressing limb regeneration in amniotes. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 328(6), pp.493–514. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
