Mixing with native broadleaf trees modified soil microbial communities of Cunninghamia lanceolata monocultures in South China
- PMID: 38505544
- PMCID: PMC10949948
- DOI: 10.3389/fmicb.2024.1372128
Mixing with native broadleaf trees modified soil microbial communities of Cunninghamia lanceolata monocultures in South China
Abstract
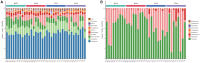
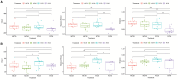
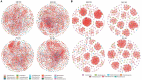
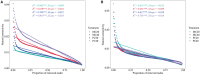
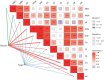
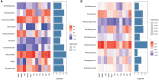
Mixing with different broadleaf trees into the monocultures of Cunninghamia lanceolata is widely adopted as an efficient transformation of the pure C. lanceolata forest. However, it is unclear how native broad-leaved trees influence the belowground ecological environment of the pure C. lanceolata culture plantation in nutrient-poor soil of South China. Herein, we aimed to investigate how a long-time mixing with native broadleaf trees shape soil microbial community of the pure C. lanceolata forest across different soil depth (0-20 cm and 20-40 cm) and to clarify relationships between the modified soil microbial community and those affected soil chemical properties. Using high-throughput sequencing technology, microbial compositions from the mixed C. lanceolata-broadleaf forest and the pure C. lanceolata forest were analyzed. Network analysis was utilized to investigate correlations among microorganisms, and network robustness was assessed by calculating network natural connectivity. Results demonstrated that the content of soil microbial biomass carbon and nitrogen, total phosphorus and pH in mixed forest stand were significantly higher than those in pure forest stand, except for available phosphorus in topsoil (0-20 cm). Simultaneously, the mixed C. lanceolata-broadleaf forest has a more homogeneous bacterial and fungal communities across different soil depth compared with the pure C. lanceolata forest, wherein the mixed forest recruited more diverse bacterial community in subsoil (20-40 cm) and reduced the diversity of fungal community in topsoil. Meanwhile, the mixed forest showed higher bacterial community stability while the pure forest showed higher fungal community stability. Moreover, bacterial communities showed significant correlations with various soil chemical indicators, whereas fungal communities exhibited correlations with only TP and pH. Therefore, the mixed C. lanceolata-broadleaf forest rely on their recruiting bacterial community to enhance and maintain the higher nutrient status of soil while the pure C. lanceolata forest rely on some specific fungi to satisfy their phosphorus requirement for survive strategy.
Keywords: microbial community structure; microbial symbiotic network; mixed Cunninghamia lanceolata-broadleaf forest; pure Cunninghamia lanceolata forest; soil microbial diversity.
Copyright © 2024 Zheng, Gu, Lu, Yang, Shuai, Li and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Response of Rhizosphere Bacterial Communities to Near-Natural Forest Management and Tree Species within Chinese Fir Plantations.Microbiol Spectr. 2023 Feb 14;11(1):e0232822. doi: 10.1128/spectrum.02328-22. Epub 2023 Jan 23. Microbiol Spectr. 2023. PMID: 36688690 Free PMC article.
-
[Characteristics of soil microbial biomass carbon and nitrogen and their relationships with soil nutrients in Cunninghamia lanceolata plantations].Ying Yong Sheng Tai Xue Bao. 2006 Dec;17(12):2292-6. Ying Yong Sheng Tai Xue Bao. 2006. PMID: 17330467 Chinese.
-
[Effects of replacing natural secondary broad-leaved forest with Cunninghamia lanceolata plantation on soil biological activities].Ying Yong Sheng Tai Xue Bao. 2005 Aug;16(8):1411-6. Ying Yong Sheng Tai Xue Bao. 2005. PMID: 16262050 Chinese.
-
[Effects of broadleaf plantation and Chinese fir (Cunninghamia lanceolata) plantation on soil carbon and nitrogen pools].Ying Yong Sheng Tai Xue Bao. 2013 Feb;24(2):345-50. Ying Yong Sheng Tai Xue Bao. 2013. PMID: 23705377 Chinese.
-
Differential response of soil microbial and animal communities along the chronosequence of Cunninghamia lanceolata at different soil depth levels in subtropical forest ecosystem.J Adv Res. 2021 Aug 11;38:41-54. doi: 10.1016/j.jare.2021.08.005. eCollection 2022 May. J Adv Res. 2021. PMID: 35572399 Free PMC article.
Cited by
-
Effects of Near-Natural Forest Management on Soil Microbial Communities in the Temperate-Subtropical Transition Zone of China.Microorganisms. 2025 Aug 15;13(8):1906. doi: 10.3390/microorganisms13081906. Microorganisms. 2025. PMID: 40871410 Free PMC article.
References
-
- Baliyarsingh B., Dash B., Nayak S., Nayak S. K. (2022). “Soil verrucomicrobia and their role in sustainable agriculture,” in Advances in Agricultural and Industrial Microbiology, Volume 1, eds S. K. Nayak, B. Baliyarsingh, I. Mannazzu, A. Singh, and B. B. Mishra (Singapore: Springer Nature Singapore), 105–124. 10.1007/978-981-16-8918-5_6 - DOI
-
- Basak B. B. (2019). Waste mica as alternative source of plant-available potassium: evaluation of agronomic potential through chemical and biological methods. Nat. Resour. Res. 28, 953–965. 10.1007/s11053-018-9430-3 - DOI
-
- Berg G., Zachow C., Müller H., Philipps J., Tilcher R. (2013). Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy 3, 648–656. 10.3390/agronomy3040648 - DOI
LinkOut - more resources
Full Text Sources

