Dietary fiber is a critical determinant of pathologic ILC2 responses and intestinal inflammation
- PMID: 38506708
- PMCID: PMC10955042
- DOI: 10.1084/jem.20232148
Dietary fiber is a critical determinant of pathologic ILC2 responses and intestinal inflammation
Abstract
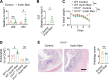
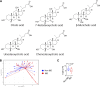
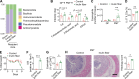
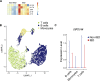
Innate lymphoid cells (ILCs) can promote host defense, chronic inflammation, or tissue protection and are regulated by cytokines and neuropeptides. However, their regulation by diet and microbiota-derived signals remains unclear. We show that an inulin fiber diet promotes Tph1-expressing inflammatory ILC2s (ILC2INFLAM) in the colon, which produce IL-5 but not tissue-protective amphiregulin (AREG), resulting in the accumulation of eosinophils. This exacerbates inflammation in a murine model of intestinal damage and inflammation in an ILC2- and eosinophil-dependent manner. Mechanistically, the inulin fiber diet elevated microbiota-derived bile acids, including cholic acid (CA) that induced expression of ILC2-activating IL-33. In IBD patients, bile acids, their receptor farnesoid X receptor (FXR), IL-33, and eosinophils were all upregulated compared with controls, implicating this diet-microbiota-ILC2 axis in human IBD pathogenesis. Together, these data reveal that dietary fiber-induced changes in microbial metabolites operate as a rheostat that governs protective versus pathologic ILC2 responses with relevance to precision nutrition for inflammatory diseases.
© 2024 Arifuzzaman et al.
Conflict of interest statement
Disclosures: C.-J. Guo reported grants from NIH and the Kenneth Rainin Foundation during the conduct of the study (NIH grants 1DP2HD101401-01, DK135816-01, the Kenneth Rainin Foundation). F.C. Schroeder reported personal fees from Ascribe Bioscience and other from Holoclara outside the submitted work. D. Artis reported personal fees from Pfizer and the Rainin Foundation outside the submitted work. No other disclosures were reported.
Figures







References
-
- Ahrens, R., Waddell A., Seidu L., Blanchard C., Carey R., Forbes E., Lampinen M., Wilson T., Cohen E., Stringer K., et al. 2008. Intestinal macrophage/epithelial cell-derived CCL11/eotaxin-1 mediates eosinophil recruitment and function in pediatric ulcerative colitis. J. Immunol. 181:7390–7399. 10.4049/jimmunol.181.10.7390 - DOI - PMC - PubMed
-
- Armstrong, H.K., Bording-Jorgensen M., Santer D.M., Zhang Z., Valcheva R., Rieger A.M., Sung-Ho Kim J., Dijk S.I., Mahmood R., Ogungbola O., et al. 2022. Unfermented beta-fructan fibers fuel inflammation in select inflammatory bowel disease patients. Gastroenterology. 164:228–240. 10.1053/j.gastro.2022.09.034 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- R01 AI095466/AI/NIAID NIH HHS/United States
- K99AI173660/NH/NIH HHS/United States
- DP2 HD101401/HD/NICHD NIH HHS/United States
- R01 DK132244/DK/NIDDK NIH HHS/United States
- R01 DK126871/DK/NIDDK NIH HHS/United States
- K99 AI173660/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 AI151599/AI/NIAID NIH HHS/United States
- R01 AI172027/AI/NIAID NIH HHS/United States
- T32 DK116970/DK/NIDDK NIH HHS/United States
- R01 AR070116/AR/NIAMS NIH HHS/United States
- R35 GM131877/GM/NIGMS NIH HHS/United States
- U01 AI095608/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

