Proteomic analysis of P. gingivalis-Lipopolysaccharide induced neuroinflammation in SH-SY5Y and HMC3 cells
- PMID: 38507186
- PMCID: PMC11336124
- DOI: 10.1007/s11357-024-01117-z
Proteomic analysis of P. gingivalis-Lipopolysaccharide induced neuroinflammation in SH-SY5Y and HMC3 cells
Abstract
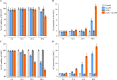
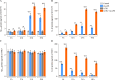
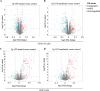
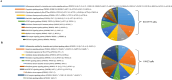
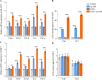
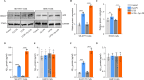
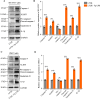
Chronic periodontitis and its keystone pathogen, Porphyromonas gingivalis, have increasingly been linked with Alzheimer's disease (AD). However, P.gingivalis-lipopolysaccharide (LPS) mediated release of neuroinflammatory proteins contributes to AD remains underexplored. In this study, we utilized data-independent acquisition mass spectrometry to characterize P.gingivalis-LPS induced profile of differentially expressed proteins associated with the neuroinflammatory response in human neuroblastoma (SH-SY5Y) and human microglial (HMC3) cells. We reported a set of 124 proteins in SH-SY5Y cells and 96 proteins in HMC3 cells whose levels were significantly upregulated or downregulated by exposure to P. gingivalis-LPS. Our findings demonstrate that P. gingivalis-LPS contributed to the elevated expressions of dementia biomarkers and pro-inflammatory cytokines that include APP, Aβ1-42, Aβ1-40, T-Tau, p-Tau, VEGF, TGF-β, IL-1β, IL-6 and TNF-α through 2 distinct pathways of extracellular sensing by cell surface receptors and intracellular cytosolic receptors. Interestingly, intracellular signaling proteins activated with P. gingivalis-LPS transfection using Lipofectamine™ 2000 had significantly higher fold change protein expression compared to the extracellular signaling with P. gingivalis-LPS treatment. Additionally, we also explored P. gingivalis-LPS mediated activation of caspase-4 dependent non canonical inflammasome pathway in both SH-SY5Y and HMC3 cells. In summary, P. gingivalis-LPS induced neuroinflammatory protein expression in SH-SY5Y and HMC3 cells, provided insights into the specific inflammatory pathways underlying the potential link between P. gingivalis-LPS infection and the pathogenesis of Alzheimer's disease and related dementias.
Keywords: Porphyromonas gingivalis; Amyloid precursor protein; Data-independent acquisition mass spectrometry; Lipopolysaccharide; Neuroinflammation; Tau protein.
© 2024. The Author(s), under exclusive licence to American Aging Association.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Bascones Martínez A, Figuero Ruiz E. Periodontal diseases as bacterial infection. Avances en Periodoncia. 2005;17. 10.4321/S1699-65852005000300002.
-
- Liu S, Butler CA, Ayton S, Reynolds EC, Dashper SG. Porphyromonas gingivalis and the pathogenesis of Alzheimer’s disease. Crit Rev Microbiol. 2023;1–11. 10.1080/1040841X.2022.2163613. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

