SUMOylation controls Hu antigen R posttranscriptional activity in liver cancer
- PMID: 38507413
- PMCID: PMC11025316
- DOI: 10.1016/j.celrep.2024.113924
SUMOylation controls Hu antigen R posttranscriptional activity in liver cancer
Abstract
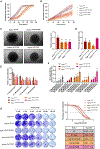
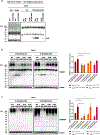
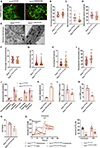
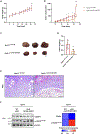
The posttranslational modification of proteins critically influences many biological processes and is a key mechanism that regulates the function of the RNA-binding protein Hu antigen R (HuR), a hub in liver cancer. Here, we show that HuR is SUMOylated in the tumor sections of patients with hepatocellular carcinoma in contrast to the surrounding tissue, as well as in human cell line and mouse models of the disease. SUMOylation of HuR promotes major cancer hallmarks, namely proliferation and invasion, whereas the absence of HuR SUMOylation results in a senescent phenotype with dysfunctional mitochondria and endoplasmic reticulum. Mechanistically, SUMOylation induces a structural rearrangement of the RNA recognition motifs that modulates HuR binding affinity to its target RNAs, further modifying the transcriptomic profile toward hepatic tumor progression. Overall, SUMOylation constitutes a mechanism of HuR regulation that could be potentially exploited as a therapeutic strategy for liver cancer.
Keywords: CP: Cancer; CP: Molecular biology; ELAVL1; HCC; PTMs; SUMO; senescence.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

