DNAJB1-PRKACA fusion neoantigens elicit rare endogenous T cell responses that potentiate cell therapy for fibrolamellar carcinoma
- PMID: 38508137
- PMCID: PMC10983114
- DOI: 10.1016/j.xcrm.2024.101469
DNAJB1-PRKACA fusion neoantigens elicit rare endogenous T cell responses that potentiate cell therapy for fibrolamellar carcinoma
Abstract
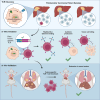
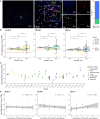
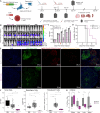
Fibrolamellar carcinoma (FLC) is a liver tumor with a high mortality burden and few treatment options. A promising therapeutic vulnerability in FLC is its driver mutation, a conserved DNAJB1-PRKACA gene fusion that could be an ideal target neoantigen for immunotherapy. In this study, we aim to define endogenous CD8 T cell responses to this fusion in FLC patients and evaluate fusion-specific T cell receptors (TCRs) for use in cellular immunotherapies. We observe that fusion-specific CD8 T cells are rare and that FLC patient TCR repertoires lack large clusters of related TCR sequences characteristic of potent antigen-specific responses, potentially explaining why endogenous immune responses are insufficient to clear FLC tumors. Nevertheless, we define two functional fusion-specific TCRs, one of which has strong anti-tumor activity in vivo. Together, our results provide insights into the fragmented nature of neoantigen-specific repertoires in humans and indicate routes for clinical development of successful immunotherapies for FLC.
Keywords: CD8 T cell; DNAJB1-PRKACA; T cell receptor; TCR repertoire; cell therapy; fibrolamellar carcinoma; gene fusion; immunotherapy; neoantigen.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.C.C., A.M.K., A.E.Z., S.E.S., and P.G.T. have a patent application for TCRs for treating FLC. J.C.C. has additional patent applications in the field of immunotherapy. A.W.P. is a member of the scientific advisory board (SAB) of Bioinformatics Solutions Inc. (Canada), shareholder and SAB member of Evaxion Biotech (Denmark), consultant for Grey Wolf Therapeutics (UK), and cofounder of Resseptor Therapeutics (Melbourne). S.G. is a co-inventor on patent applications in the fields of cell/gene therapy for cancer, consultant of TESSA Therapeutics, member of the Data and Safety Monitoring Board of Immatics, and SAB member of Be Biopharma and has received honoraria from Tidal, Catamaran Bio, and Sanofi within the last 2 years. S.E.S. is a cofounder, stockholder, and paid consultant for Gliknik Inc., serves on the SAB and holds stock options for Virion Inc., and receives royalties from the Mayo Clinic for licensed IP surrounding manipulation of the PD-1:PD-L1 pathway for cancer treatment. P.G.T. is on the SAB of Immunoscape and Shennon Bio, received personal fees and research support from Elevate Bio, and consulted for 10×, Illumina, Pfizer, Cytoagents, and JNJ.
Figures




References
-
- Dinh T.A., Utria A.F., Barry K.C., Ma R., Abou-Alfa G.K., Gordan J.D., Jaffee E.M., Scott J.D., Zucman-Rossi J., O’Neill A.F., et al. A framework for fibrolamellar carcinoma research and clinical trials. Nat. Rev. Gastroenterol. Hepatol. 2022;19:328–342. doi: 10.1038/s41575-022-00580-3. - DOI - PMC - PubMed
-
- Eggert T., McGlynn K.A., Duffy A., Manns M.P., Greten T.F., Altekruse S.F. Fibrolamellar hepatocellular carcinoma in the USA, 2000-2010: A detailed report on frequency, treatment and outcome based on the Surveillance, Epidemiology, and End Results database. United Eur. Gastroenterol. J. 2013;1:351–357. doi: 10.1177/2050640613501507. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

