DNAJB8 oligomerization is mediated by an aromatic-rich motif that is dispensable for substrate activity
- PMID: 38508190
- PMCID: PMC11162344
- DOI: 10.1016/j.str.2024.02.015
DNAJB8 oligomerization is mediated by an aromatic-rich motif that is dispensable for substrate activity
Abstract
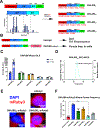
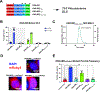
J-domain protein (JDP) molecular chaperones have emerged as central players that maintain a healthy proteome. The diverse members of the JDP family function as monomers/dimers and a small subset assemble into micron-sized oligomers. The oligomeric JDP members have eluded structural characterization due to their low-complexity, intrinsically disordered middle domains. This in turn, obscures the biological significance of these larger oligomers in protein folding processes. Here, we identified a short, aromatic motif within DNAJB8 that drives self-assembly through π-π stacking and determined its X-ray structure. We show that mutations in the motif disrupt DNAJB8 oligomerization in vitro and in cells. DNAJB8 variants that are unable to assemble bind to misfolded tau seeds more specifically and retain capacity to reduce protein aggregation in vitro and in cells. We propose a new model for DNAJB8 function in which the sequences in the low-complexity domains play distinct roles in assembly and substrate activity.
Keywords: Alzheimer‘s disease; DNAJB6; DNAJB8; J-domain proteins; LARKS; chaperone; neurodegeneration; polyQ; proteostasis; tau.
Copyright © 2024 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Update of
-
DNAJB8 oligomerization is mediated by an aromatic-rich motif that is dispensable for substrate activity.bioRxiv [Preprint]. 2024 Jan 23:2023.03.06.531355. doi: 10.1101/2023.03.06.531355. bioRxiv. 2024. Update in: Structure. 2024 Jun 6;32(6):662-678.e8. doi: 10.1016/j.str.2024.02.015. PMID: 36945632 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

