Circular RNA as a source of neoantigens for cancer vaccines
- PMID: 38508656
- PMCID: PMC10952939
- DOI: 10.1136/jitc-2023-008402
Circular RNA as a source of neoantigens for cancer vaccines
Abstract
Background: The effectiveness of somatic neoantigen-based immunotherapy is often hindered by the limited number of mutations in tumors with low to moderate mutation burden. Focusing on microsatellite-stable colorectal cancer (CRC), this study investigates the potential of tumor-associated circular RNAs (circRNAs) as an alternative source of neoepitopes in CRC.
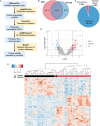
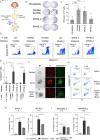
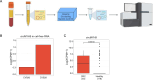
Methods: Tumor-associated circRNAs in CRC were identified using the MiOncoCirc database and ribo-depletion RNA sequencing of paired clinical normal and tumor samples. Candidate circRNA expression was validated by quantitative real-time PCR (RT-qPCR) using divergent primers. TransCirc database was used for translation prediction. Human leukocyte antigen binding affinity of open reading frames from potentially translatable circRNA was predicted using pVACtools. Strong binders from messenger RNA-encoded proteins were excluded using BlastP. The immunogenicity of the candidate antigens was functionally validated through stimulation of naïve CD8+ T cells against the predicted neoepitopes and subsequent analysis of the T cells through enzyme-linked immunospot (ELISpot) assay, intracellular cytokine staining (ICS) and granzyme B (GZMB) reporter. The cytotoxicity of T cells trained with antigen peptides was further tested using patient-derived organoids.
Results: We identified a neoepitope from circRAPGEF5 that is upregulated in CRC tumor samples from MiOncoCirc database, and two neoepitopes from circMYH9, which is upregulated across various tumor samples from our matched clinical samples. The translation potential of candidate peptides was supported by Clinical Proteomic Tumor Analysis Consortium database using PepQuery. The candidate peptides elicited antigen-specific T cells response and expansion, evidenced by various assays including ELISpot, ICS and GZMB reporter. Furthermore, T cells trained with circMYH9 peptides were able to specifically target and eliminate tumor-derived organoids but not match normal organoids. This observation underscores the potential of circRNAs as a source of immunogenic neoantigens. Lastly, circMYH9 was enriched in the liquid biopsies of patients with CRC, thus enabling a detection-to-vaccination treatment strategy for patients with CRC.
Conclusions: Our findings underscore the feasibility of tumor-associated circRNAs as an alternative source of neoantigens for cancer vaccines targeting tumors with moderate mutation levels.
Keywords: circular RNA; colorectal cancer; neoantigen.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: No, there are no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
