Reverse-engineering the anti-MUC1 antibody 139H2 by mass spectrometry-based de novo sequencing
- PMID: 38508723
- PMCID: PMC10955041
- DOI: 10.26508/lsa.202302366
Reverse-engineering the anti-MUC1 antibody 139H2 by mass spectrometry-based de novo sequencing
Abstract
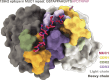
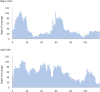
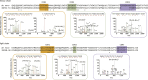
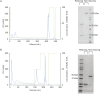
Mucin 1 (MUC1) is a transmembrane mucin expressed at the apical surface of epithelial cells at mucosal surfaces. MUC1 has a barrier function against bacterial invasion and is well known for its aberrant expression and glycosylation in adenocarcinomas. The MUC1 extracellular domain contains a variable number of tandem repeats (VNTR) of 20 amino acids, which are heavily O-linked glycosylated. Monoclonal antibodies against the MUC1 VNTR are powerful research tools with applications in the diagnosis and treatment of MUC1-expressing cancers. Here, we report direct mass spectrometry-based sequencing of anti-MUC1 hybridoma-derived 139H2 IgG, enabling reverse-engineering of the functional recombinant monoclonal antibody. The crystal structure of the 139H2 Fab fragment in complex with the MUC1 epitope was solved, revealing the molecular basis of 139H2 binding specificity to MUC1 and its tolerance to O-glycosylation of the VNTR. The available sequence of 139H2 will allow further development of MUC1-related diagnostic, targeting, and treatment strategies.
© 2024 Peng et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





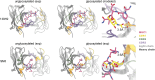
Similar articles
-
Heterogeneity of mucin gene expression in normal and neoplastic tissues.Cancer Res. 1993 Feb 1;53(3):641-51. Cancer Res. 1993. PMID: 7678777
-
Bovine Muc1 is a highly polymorphic gene encoding an extensively glycosylated mucin that binds bacteria.J Dairy Sci. 2009 Oct;92(10):5276-91. doi: 10.3168/jds.2009-2216. J Dairy Sci. 2009. PMID: 19762846
-
Characterization of a new MUC1 monoclonal antibody (VU-2-G7) directed to the glycosylated PDTR sequence of MUC1.Tumour Biol. 2000 Jul-Aug;21(4):197-210. doi: 10.1159/000030126. Tumour Biol. 2000. PMID: 10867613
-
MUC1: a target molecule for cancer therapy.Cancer Biol Ther. 2007 Apr;6(4):481-6. doi: 10.4161/cbt.6.4.4201. Cancer Biol Ther. 2007. PMID: 18027437 Review.
-
Expression of mucin antigens in human cancers and its relationship with malignancy potential.Pathol Int. 1997 Dec;47(12):813-30. doi: 10.1111/j.1440-1827.1997.tb03713.x. Pathol Int. 1997. PMID: 9503463 Review.
Cited by
-
Direct Mass Spectrometry-Based Detection and Antibody Sequencing of Monoclonal Gammopathy of Undetermined Significance from Patient Serum: A Case Study.J Proteome Res. 2023 Sep 1;22(9):3022-3028. doi: 10.1021/acs.jproteome.3c00330. Epub 2023 Jul 27. J Proteome Res. 2023. PMID: 37499263 Free PMC article.
-
Structural Basis for Postfusion-Specific Binding to the Respiratory Syncytial Virus F Protein by the Canonical Antigenic Site I Antibody 131-2a.ACS Infect Dis. 2025 Aug 8;11(8):2357-2366. doi: 10.1021/acsinfecdis.5c00368. Epub 2025 Jul 22. ACS Infect Dis. 2025. PMID: 40693554 Free PMC article.
-
Simultaneous polyclonal antibody sequencing and epitope mapping by cryo electron microscopy and mass spectrometry.Elife. 2025 Apr 23;14:RP101322. doi: 10.7554/eLife.101322. Elife. 2025. PMID: 40266252 Free PMC article.
-
A Handle on Mass Coincidence Errors in De Novo Sequencing of Antibodies by Bottom-up Proteomics.J Proteome Res. 2024 Aug 2;23(8):3552-3559. doi: 10.1021/acs.jproteome.4c00188. Epub 2024 Jun 27. J Proteome Res. 2024. PMID: 38932690 Free PMC article.
References
-
- Frey A, Giannasca KT, Weltzin R, Giannasca PJ, Reggio H, Lencer WI, Neutra MR (1996) Role of the glycocalyx in regulating access of microparticles to apical plasma membranes of intestinal epithelial cells: Implications for microbial attachment and oral vaccine targeting. J Exp Med 184: 1045–1059. 10.1084/jem.184.3.1045 - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
