The role of host traits and geography in shaping the gut microbiome of insectivorous bats
- PMID: 38509042
- PMCID: PMC11036801
- DOI: 10.1128/msphere.00087-24
The role of host traits and geography in shaping the gut microbiome of insectivorous bats
Abstract
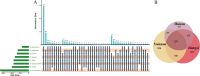
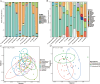
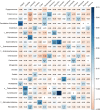
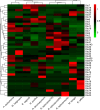
The gut microbiome is a symbiotic microbial community associated with the host and plays multiple important roles in host physiology, nutrition, and health. A number of factors have been shown to influence the gut microbiome, among which diet is considered to be one of the most important; however, the relationship between diet composition and gut microbiota in wild mammals is still not well recognized. Herein, we characterized the gut microbiota of bats and examined the effects of diet, host taxa, body size, gender, elevation, and latitude on the gut microbiota. The cytochrome C oxidase subunit I (COI) gene and 16S rRNA gene amplicons were sequenced from the feces of eight insectivorous bat species in southern China, including Miniopterus fuliginosus, Aselliscus stoliczkanus, Myotis laniger, Rhinolophus episcopus, Rhinolophus osgoodi, Rhinolophus ferrumequinum, Rhinolophus affinis, and Rhinolophus pusillus. The results showed that the composition of gut microbiome and diet exhibited significant differences among bat species. Diet composition and gut microbiota were significantly correlated at the order, family, genus, and operational taxonomic unit levels, while certain insects had a marked effect on the gut microbiome at specific taxonomic levels. In addition, elevation, latitude, body weight of bats, and host species had significant effects on the gut microbiome, but phylosymbiosis between host phylogeny and gut microbiome was lacking. These findings clarify the relationship between gut microbiome and diet and contribute to improving our understanding of host ecology and the evolution of the gut microbiome in wild mammals.
Importance: The gut microbiome is critical for the adaptation of wildlife to the dynamic environment. Bats are the second-largest group of mammals with short intestinal tract, yet their gut microbiome is still poorly studied. Herein, we explored the relationships between gut microbiome and food composition, host taxa, body size, gender, elevation, and latitude. We found a significant association between diet composition and gut microbiome in insectivorous bats, with certain insect species having major impacts on gut microbiome. Factors like species taxa, body weight, elevation, and latitude also affected the gut microbiome, but we failed to detect phylosymbiosis between the host phylogeny and the gut microbiome. Overall, our study presents novel insights into how multiple factors shape the bat's gut microbiome together and provides a study case on host-microbe interactions in wildlife.
Keywords: 16S rRNA; diet composition; gut microbiome; high-throughput sequencing; insectivorous bats.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Amato KR. 2013. Co-evolution in context: the importance of studying gut microbiomes in wild animals. Microbiome Sci Med 1:10–29. doi: 10.2478/micsm-2013-0002 - DOI
Publication types
MeSH terms
Substances
Grants and funding
- 32171525/MOST | National Natural Science Foundation of China (NSFC)
- 31961123001/MOST | National Natural Science Foundation of China (NSFC)
- 20220101291JC/| Natural Science Foundation of Jilin Province (Jilin Natural Science Foundation)
- 20230101160JC/| Natural Science Foundation of Jilin Province (Jilin Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Medical

