Metabolic targeting of cancer associated fibroblasts overcomes T-cell exclusion and chemoresistance in soft-tissue sarcomas
- PMID: 38509063
- PMCID: PMC10954767
- DOI: 10.1038/s41467-024-46504-4
Metabolic targeting of cancer associated fibroblasts overcomes T-cell exclusion and chemoresistance in soft-tissue sarcomas
Abstract
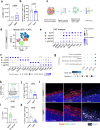
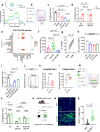
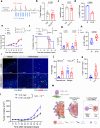
T cell-based immunotherapies have exhibited promising outcomes in tumor control; however, their efficacy is limited in immune-excluded tumors. Cancer-associated fibroblasts (CAFs) play a pivotal role in shaping the tumor microenvironment and modulating immune infiltration. Despite the identification of distinct CAF subtypes using single-cell RNA-sequencing (scRNA-seq), their functional impact on hindering T-cell infiltration remains unclear, particularly in soft-tissue sarcomas (STS) characterized by low response rates to T cell-based therapies. In this study, we characterize the STS microenvironment using murine models (in female mice) with distinct immune composition by scRNA-seq, and identify a subset of CAFs we termed glycolytic cancer-associated fibroblasts (glyCAF). GlyCAF rely on GLUT1-dependent expression of CXCL16 to impede cytotoxic T-cell infiltration into the tumor parenchyma. Targeting glycolysis decreases T-cell restrictive glyCAF accumulation at the tumor margin, thereby enhancing T-cell infiltration and augmenting the efficacy of chemotherapy. These findings highlight avenues for combinatorial therapeutic interventions in sarcomas and possibly other solid tumors. Further investigations and clinical trials are needed to validate these potential strategies and translate them into clinical practice.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
- R01 CA258265/CA/NCI NIH HHS/United States
- SFA 15-19/Sarcoma Foundation of America (Sarcoma Foundation of America, Inc.)
- CA258265/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R00 HL141702/HL/NHLBI NIH HHS/United States
- CA212200/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

