Lactobacillus rhamnosus GG ameliorates hyperuricemia in a novel model
- PMID: 38509085
- PMCID: PMC10954633
- DOI: 10.1038/s41522-024-00486-9
Lactobacillus rhamnosus GG ameliorates hyperuricemia in a novel model
Abstract
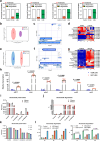
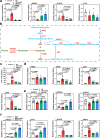
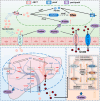
Hyperuricemia (HUA) is a metabolic syndrome caused by abnormal purine metabolism. Although recent studies have noted a relationship between the gut microbiota and gout, whether the microbiota could ameliorate HUA-associated systemic purine metabolism remains unclear. In this study, we constructed a novel model of HUA in geese and investigated the mechanism by which Lactobacillus rhamnosus GG (LGG) could have beneficial effects on HUA. The administration of antibiotics and fecal microbiota transplantation (FMT) experiments were used in this HUA goose model. The effects of LGG and its metabolites on HUA were evaluated in vivo and in vitro. Heterogeneous expression and gene knockout of LGG revealed the mechanism of LGG. Multi-omics analysis revealed that the Lactobacillus genus is associated with changes in purine metabolism in HUA. This study showed that LGG and its metabolites could alleviate HUA through the gut-liver-kidney axis. Whole-genome analysis, heterogeneous expression, and gene knockout of LGG enzymes ABC-type multidrug transport system (ABCT), inosine-uridine nucleoside N-ribohydrolase (iunH), and xanthine permease (pbuX) demonstrated the function of nucleoside degradation in LGG. Multi-omics and a correlation analysis in HUA patients and this goose model revealed that a serum proline deficiency, as well as changes in Collinsella and Lactobacillus, may be associated with the occurrence of HUA. Our findings demonstrated the potential of a goose model of diet-induced HUA, and LGG and proline could be promising therapies for HUA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

