RNA-binding proteins potentially regulate the alternative splicing of cell cycle-associated genes in proliferative diabetic retinopathy
- PMID: 38509306
- PMCID: PMC10954754
- DOI: 10.1038/s41598-024-57516-x
RNA-binding proteins potentially regulate the alternative splicing of cell cycle-associated genes in proliferative diabetic retinopathy
Abstract
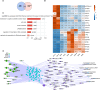
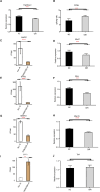
RNA-binding proteins (RBPs) contribute to the pathogenesis of proliferative diabetic retinopathy (PDR) by regulating gene expression through alternative splicing events (ASEs). However, the RBPs differentially expressed in PDR and the underlying mechanisms remain unclear. Thus, this study aimed to identify the differentially expressed genes in the neovascular membranes (NVM) and retinas of patients with PDR. The public transcriptome dataset GSE102485 was downloaded from the Gene Expression Omnibus database, and samples of PDR and normal retinas were analyzed. A mouse model of oxygen-induced retinopathy was used to confirm the results. The top 20 RBPs were screened for co-expression with alternative splicing genes (ASGs). A total of 403 RBPs were abnormally expressed in the NVM and retina samples. Functional analysis demonstrated that the ASGs were enriched in cell cycle pathways. Cell cycle-associated ASEs and an RBP-AS regulatory network, including 15 RBPs and their regulated ASGs, were extracted. Splicing factor proline/glutamine rich (SFPQ), microtubule-associated protein 1 B (MAP1B), heat-shock protein 90-alpha (HSP90AA1), microtubule-actin crosslinking factor 1 (MACF1), and CyclinH (CCNH) expression remarkably differed in the mouse model. This study provides novel insights into the RBP-AS interaction network in PDR and for developing screening and treatment options to prevent diabetic retinopathy-related blindness.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
RNA-Binding Proteins and Alternative Splicing Genes Are Coregulated in Human Retinal Endothelial Cells Treated with High Glucose.J Diabetes Res. 2022 Mar 9;2022:7680513. doi: 10.1155/2022/7680513. eCollection 2022. J Diabetes Res. 2022. PMID: 35308095 Free PMC article.
-
Upregulation of HMOX1 associated with M2 macrophage infiltration and ferroptosis in proliferative diabetic retinopathy.Int Immunopharmacol. 2024 Jun 15;134:112231. doi: 10.1016/j.intimp.2024.112231. Epub 2024 May 12. Int Immunopharmacol. 2024. PMID: 38739977
-
Regulation of Alternative Splicing of Lipid Metabolism Genes in Sepsis-Induced Liver Damage by RNA-Binding Proteins.Inflammation. 2024 Dec;47(6):1952-1968. doi: 10.1007/s10753-024-02017-2. Epub 2024 May 9. Inflammation. 2024. PMID: 38727856 Free PMC article.
-
Advances in the study of RNA-binding proteins in diabetic complications.Mol Metab. 2022 Aug;62:101515. doi: 10.1016/j.molmet.2022.101515. Epub 2022 May 18. Mol Metab. 2022. PMID: 35597446 Free PMC article. Review.
-
Prognostic factors for the development and progression of proliferative diabetic retinopathy in people with diabetic retinopathy.Cochrane Database Syst Rev. 2023 Feb 22;2(2):CD013775. doi: 10.1002/14651858.CD013775.pub2. Cochrane Database Syst Rev. 2023. PMID: 36815723 Free PMC article. Review.
Cited by
-
A Multifaceted Giant Protein Microtubule-Actin Cross-Linking Factor 1.Int J Mol Sci. 2025 Mar 30;26(7):3204. doi: 10.3390/ijms26073204. Int J Mol Sci. 2025. PMID: 40244019 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

