Transcription-replication conflicts underlie sensitivity to PARP inhibitors
- PMID: 38509368
- PMCID: PMC11006605
- DOI: 10.1038/s41586-024-07217-2
Transcription-replication conflicts underlie sensitivity to PARP inhibitors
Abstract
An important advance in cancer therapy has been the development of poly(ADP-ribose) polymerase (PARP) inhibitors for the treatment of homologous recombination (HR)-deficient cancers1-6. PARP inhibitors trap PARPs on DNA. The trapped PARPs are thought to block replisome progression, leading to formation of DNA double-strand breaks that require HR for repair7. Here we show that PARP1 functions together with TIMELESS and TIPIN to protect the replisome in early S phase from transcription-replication conflicts. Furthermore, the synthetic lethality of PARP inhibitors with HR deficiency is due to an inability to repair DNA damage caused by transcription-replication conflicts, rather than by trapped PARPs. Along these lines, inhibiting transcription elongation in early S phase rendered HR-deficient cells resistant to PARP inhibitors and depleting PARP1 by small-interfering RNA was synthetic lethal with HR deficiency. Thus, inhibiting PARP1 enzymatic activity may suffice for treatment efficacy in HR-deficient settings.
© 2024. The Author(s).
Conflict of interest statement
T.D.H. and S.K.S. are founders and stockholders of FoRx Therapeutics. S.K.S., G.G.R., A.F. and L.G.I. are employees of FoRx Therapeutics. The other authors declare no competing interests.
Figures








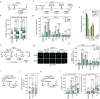
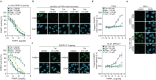
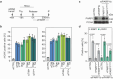
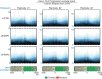


References
-
- Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

