The homeobox transcription factor DUXBL controls exit from totipotency
- PMID: 38509386
- PMCID: PMC11149696
- DOI: 10.1038/s41588-024-01692-z
The homeobox transcription factor DUXBL controls exit from totipotency
Abstract
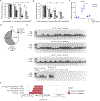
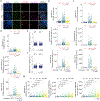
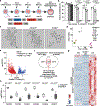
In mice, exit from the totipotent two-cell (2C) stage embryo requires silencing of the 2C-associated transcriptional program. However, the molecular mechanisms involved in this process remain poorly understood. Here we demonstrate that the 2C-specific transcription factor double homeobox protein (DUX) mediates an essential negative feedback loop by inducing the expression of DUXBL to promote this silencing. We show that DUXBL gains accessibility to DUX-bound regions specifically upon DUX expression. Furthermore, we determine that DUXBL interacts with TRIM24 and TRIM33, members of the TRIM superfamily involved in gene silencing, and colocalizes with them in nuclear foci upon DUX expression. Importantly, DUXBL overexpression impairs 2C-associated transcription, whereas Duxbl inactivation in mouse embryonic stem cells increases DUX-dependent induction of the 2C-transcriptional program. Consequently, DUXBL deficiency in embryos results in sustained expression of 2C-associated transcripts leading to early developmental arrest. Our study identifies DUXBL as an essential regulator of totipotency exit enabling the first divergence of cell fates.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing interests.
Figures














References
-
- Riveiro AR, & Brickman JM From pluripotency to totipotency: An experimentalist’s guide to cellular potency. Development (Cambridge, England) 147, dev189845 (2020). - PubMed
-
- Sha QQ, Zhang J, & Fan HY A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals. Biology of Reproduction 101, 579–590 (2019). - PubMed
-
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, Nix DA, Peterson CM, Tapscott SJ, Carrell DT. & Cairns BR. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925–934 (2017). - PMC - PubMed
METHODS-ONLY REFERENCES
-
- Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. 10.14806/ej.17.1.200. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

