Genome sequencing and analysis of black flounder (Paralichthys orbignyanus) reveals new insights into Pleuronectiformes genomic size and structure
- PMID: 38509481
- PMCID: PMC10956332
- DOI: 10.1186/s12864-024-10081-z
Genome sequencing and analysis of black flounder (Paralichthys orbignyanus) reveals new insights into Pleuronectiformes genomic size and structure
Abstract
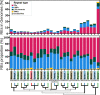
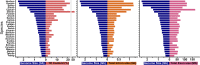
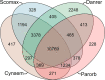
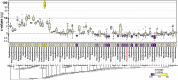
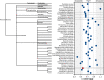
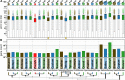
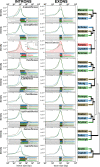
Black flounder (Paralichthys orbignyanus, Pleuronectiformes) is a commercially significant marine fish with promising aquaculture potential in Argentina. Despite extensive studies on Black flounder aquaculture, its limited genetic information available hampers the crucial role genetics plays in the development of this activity. In this study, we first employed Illumina sequencing technology to sequence the entire genome of Black flounder. Utilizing two independent libraries-one from a female and another from a male-with 150 bp paired-end reads, a mean insert length of 350 bp, and over 35 X-fold coverage, we achieved assemblies resulting in a genome size of ~ 538 Mbp. Analysis of the assemblies revealed that more than 98% of the core genes were present, with more than 78% of them having more than 50% coverage. This indicates a somehow complete and accurate genome at the coding sequence level. This genome contains 25,231 protein-coding genes, 445 tRNAs, 3 rRNAs, and more than 1,500 non-coding RNAs of other types. Black flounder, along with pufferfishes, seahorses, pipefishes, and anabantid fish, displays a smaller genome compared to most other teleost groups. In vertebrates, the number of transposable elements (TEs) is often correlated with genome size. However, it remains unclear whether the sizes of introns and exons also play a role in determining genome size. Hence, to elucidate the potential factors contributing to this reduced genome size, we conducted a comparative genomic analysis between Black flounder and other teleost orders to determine if the small genomic size could be explained by repetitive elements or gene features, including the whole genome genes and introns sizes. We show that the smaller genome size of flounders can be attributed to several factors, including changes in the number of repetitive elements, and decreased gene size, particularly due to lower amount of very large and small introns. Thus, these components appear to be involved in the genome reduction in Black flounder. Despite these insights, the full implications and potential benefits of genome reduction in Black flounder for reproduction and aquaculture remain incompletely understood, necessitating further research.
Keywords: Paralichthys orbignyanus; Black flounder; Ensembl; Evolution; Genome; Intron size; Pleuronectiformes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Nelson JS, Grande TC, Wilson MVH. Fishes of the World: Nelson/Fishes of the World. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2016.
-
- Seitz AC. Flatfishes: Biology and Exploitation. Rev Fish Biol Fish. 2008;18:249–250. doi: 10.1007/s11160-007-9072-8. - DOI
-
- Seikai T. Flounder Culture and Its Challenges in Asia. Rev Fish Sci. 2002;10:421–432. doi: 10.1080/20026491051721. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

