JCAD deficiency delayed liver regenerative repair through the Hippo-YAP signalling pathway
- PMID: 38509842
- PMCID: PMC10955226
- DOI: 10.1002/ctm2.1630
JCAD deficiency delayed liver regenerative repair through the Hippo-YAP signalling pathway
Abstract
Background and aims: Liver regeneration retardation post partial hepatectomy (PH) is a common clinical problem after liver transplantation. Identification of key regulators in liver regeneration post PH may be beneficial for clinically improving the prognosis of patients after liver transplantation. This study aimed to clarify the function of junctional protein-associated with coronary artery disease (JCAD) in liver regeneration post PH and to reveal the underlying mechanisms.
Methods: JCAD knockout (JCAD-KO), liver-specific JCAD-KO (Jcad△Hep) mice and their control group were subjected to 70% PH. RNA sequencing was conducted to unravel the related signalling pathways. Primary hepatocytes from KO mice were treated with epidermal growth factor (EGF) to evaluate DNA replication. Fluorescent ubiquitination-based cell cycle indicator (FUCCI) live-imaging system was used to visualise the phases of cell cycle.
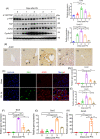
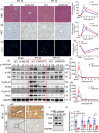
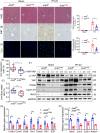
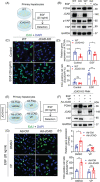
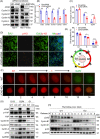
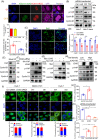
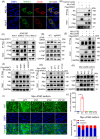
Results: Both global and liver-specific JCAD deficiency postponed liver regeneration after PH as indicated by reduced gene expression of cell cycle transition and DNA replication. Prolonged retention in G1 phase and failure to transition over the cell cycle checkpoint in JCAD-KO cell line was indicated by a FUCCI live-imaging system as well as pharmacologic blockage. JCAD replenishment by adenovirus reversed the impaired DNA synthesis in JCAD-KO primary hepatocyte in exposure to EGF, which was abrogated by a Yes-associated protein (YAP) inhibitor, verteporfin. Mechanistically, JCAD competed with large tumour suppressor 2 (LATS2) for WWC1 interaction, leading to LATS2 inhibition and thereafter YAP activation, and enhanced expression of cell cycle-associated genes.
Conclusion: JCAD deficiency led to delayed regeneration after PH as a result of blockage in cell cycle progression through the Hippo-YAP signalling pathway. These findings uncovered novel functions of JCAD and suggested a potential strategy for improving graft growth and function post liver transplantation.
Key points: JCAD deficiency leads to an impaired liver growth after PH due to cell division blockage. JCAD competes with LATS2 for WWC1 interaction, resulting in LATS2 inhibition, YAP activation and enhanced expression of cell cycle-associated genes. Delineation of JCADHippoYAP signalling pathway would facilitate to improve prognosis of acute liver failure and graft growth in living-donor liver transplantation.
Keywords: Hippo–YAP signalling pathway; JCAD; WWC1; cell cycle phase visualisation; liver regeneration.
© 2024 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare they have no competing financial interests associated with conduction of this investigation.
Figures

References
-
- Chen X, Murad M, Cui YY, et al. miRNA regulation of liver growth after 50% partial hepatectomy and small size grafts in rats. Transplantation. 2011;91(3):293‐299. - PubMed
-
- Rela M, Reddy MS. Living donor liver transplant (LDLT) is the way forward in Asia. Hepatol Int. 2017;11(2):148‐151. - PubMed
-
- Adam R, Karam V, Delvart V, et al. Evolution of indications and results of liver transplantation in Europe. A report from the European Liver Transplant Registry (ELTR). J Hepatol. 2012;57(3):675‐688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
