Cardiac transcriptomic changes induced by early CKD in mice reveal novel pathways involved in the pathogenesis of Cardiorenal syndrome type 4
- PMID: 38509984
- PMCID: PMC10950824
- DOI: 10.1016/j.heliyon.2024.e27468
Cardiac transcriptomic changes induced by early CKD in mice reveal novel pathways involved in the pathogenesis of Cardiorenal syndrome type 4
Abstract
Background: Cardiorenal syndrome (CRS) type 4 is prevalent among the chronic kidney disease (CKD) population, with many patients dying from cardiovascular complications. However, limited data regarding cardiac transcriptional changes induced early by CKD is available.
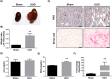
Methods: We used a murine unilateral ureteral obstruction (UUO) model to evaluate renal damage, cardiac remodeling, and transcriptional regulation at 21 days post-surgery through histological analysis, RT-qPCR, RNA-seq, and bioinformatics.
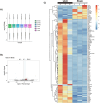
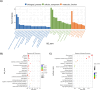
Results: UUO leads to significant kidney injury, low uremia, and pathological cardiac remodeling, evidenced by increased collagen deposition and smooth muscle alpha-actin 2 expression. RNA-seq analysis identified 76 differentially expressed genes (DEGs) in UUO hearts. Upregulated DEGs were significantly enriched in cell cycle and cell division pathways, immune responses, cardiac repair, inflammation, proliferation, oxidative stress, and apoptosis. Gene Set Enrichment Analysis further revealed mitochondrial oxidative bioenergetic pathways, autophagy, and peroxisomal pathways are downregulated in UUO hearts. Vimentin was also identified as an UUO-upregulated transcript.
Conclusions: Our results emphasize the relevance of extensive transcriptional changes, mitochondrial dysfunction, homeostasis deregulation, fatty-acid metabolism alterations, and vimentin upregulation in CRS type 4 development.
Keywords: CKD; Cardiorenal; Heart; Kidney; Transcriptomics; Uropathy; mRNA.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Pathological cardiac remodeling occurs early in CKD mice from unilateral urinary obstruction, and is attenuated by Enalapril.Sci Rep. 2018 Oct 31;8(1):16087. doi: 10.1038/s41598-018-34216-x. Sci Rep. 2018. PMID: 30382174 Free PMC article.
-
Eplerenone inhibits UUO-induced lymphangiogenesis and cardiac fibrosis by attenuating inflammatory injury.Int Immunopharmacol. 2022 Jul;108:108759. doi: 10.1016/j.intimp.2022.108759. Epub 2022 Apr 12. Int Immunopharmacol. 2022. PMID: 35428023
-
Eplerenone Prevents Cardiac Fibrosis by Inhibiting Angiogenesis in Unilateral Urinary Obstruction Rats.J Renin Angiotensin Aldosterone Syst. 2022 Sep 17;2022:1283729. doi: 10.1155/2022/1283729. eCollection 2022. J Renin Angiotensin Aldosterone Syst. 2022. PMID: 36185701 Free PMC article.
-
Redox signaling pathways in unilateral ureteral obstruction (UUO)-induced renal fibrosis.Free Radic Biol Med. 2021 Aug 20;172:65-81. doi: 10.1016/j.freeradbiomed.2021.05.034. Epub 2021 May 30. Free Radic Biol Med. 2021. PMID: 34077780 Review.
-
Mitochondrial Dysfunction: An Emerging Link in the Pathophysiology of Cardiorenal Syndrome.Front Cardiovasc Med. 2022 Feb 25;9:837270. doi: 10.3389/fcvm.2022.837270. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35282359 Free PMC article. Review.
Cited by
-
The Sigma-1 Receptor Exacerbates Cardiac Dysfunction Induced by Obstructive Nephropathy: A Role for Sexual Dimorphism.Biomedicines. 2024 Aug 20;12(8):1908. doi: 10.3390/biomedicines12081908. Biomedicines. 2024. PMID: 39200372 Free PMC article.
References
-
- Rangaswami J., Bhalla V., Blair J.E.A., Chang T.I., Costa S., Lentine K.L., Lerma E.V., Mezue K., Molitch M., Mullens W., Ronco C., Tang W.H.W., McCullough P.A., American Heart Association Council on the Kidney in Cardiovascular D, Council on Clinical C Cardiorenal syndrome: Classification, pathophysiology, diagnosis, and treatment Strategies: a scientific statement from the American heart association. Circulation. 2019;139(16):e840–e878. doi: 10.1161/CIR.0000000000000664. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

