YAP induces a neonatal-like pro-renewal niche in the adult heart
- PMID: 38510108
- PMCID: PMC10954255
- DOI: 10.1038/s44161-024-00428-w
YAP induces a neonatal-like pro-renewal niche in the adult heart
Abstract
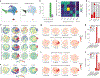
After myocardial infarction (MI), mammalian hearts do not regenerate, and the microenvironment is disrupted. Hippo signaling loss of function with activation of transcriptional co-factor YAP induces heart renewal and rebuilds the post-MI microenvironment. In this study, we investigated adult renewal-competent mouse hearts expressing an active version of YAP, called YAP5SA, in cardiomyocytes (CMs). Spatial transcriptomics and single-cell RNA sequencing revealed a conserved, renewal-competent CM cell state called adult (a)CM2 with high YAP activity. aCM2 co-localized with cardiac fibroblasts (CFs) expressing complement pathway component C3 and macrophages (MPs) expressing C3ar1 receptor to form a cellular triad in YAP5SA hearts and renewal-competent neonatal hearts. Although aCM2 was detected in adult mouse and human hearts, the cellular triad failed to co-localize in these non-renewing hearts. C3 and C3ar1 loss-of-function experiments indicated that C3a signaling between MPs and CFs was required to assemble the pro-renewal aCM2, C3+ CF and C3ar1+ MP cellular triad.
Figures



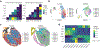













References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
