Quantitative proteome dynamics across embryogenesis in a model chordate
- PMID: 38510129
- PMCID: PMC10951915
- DOI: 10.1016/j.isci.2024.109355
Quantitative proteome dynamics across embryogenesis in a model chordate
Abstract
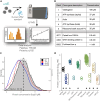
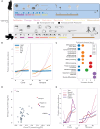
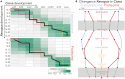
The evolution of gene expression programs underlying the development of vertebrates remains poorly characterized. Here, we present a comprehensive proteome atlas of the model chordate Ciona, covering eight developmental stages and ∼7,000 translated genes, accompanied by a multi-omics analysis of co-evolution with the vertebrate Xenopus. Quantitative proteome comparisons argue against the widely held hourglass model, based solely on transcriptomic profiles, whereby peak conservation is observed during mid-developmental stages. Our analysis reveals maximal divergence at these stages, particularly gastrulation and neurulation. Together, our work provides a valuable resource for evaluating conservation and divergence of multi-omics profiles underlying the diversification of vertebrates.
Keywords: Animals; Embryology; Evolutionary developmental biology; Proteomics; Transcriptomics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

