Signatures of disease outcome severity in the intestinal fungal and bacterial microbiome of COVID-19 patients
- PMID: 38510960
- PMCID: PMC10952111
- DOI: 10.3389/fcimb.2024.1352202
Signatures of disease outcome severity in the intestinal fungal and bacterial microbiome of COVID-19 patients
Abstract
Background: COVID-19, whose causative pathogen is the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), was declared a pandemic in March 2020. The gastrointestinal tract is one of the targets of this virus, and mounting evidence suggests that gastrointestinal symptoms may contribute to disease severity. The gut-lung axis is involved in the immune response to SARS-CoV-2; therefore, we investigated whether COVID-19 patients' bacterial and fungal gut microbiome composition was linked to disease clinical outcome.
Methods: In May 2020, we collected stool samples and patient records from 24 hospitalized patients with laboratory-confirmed SARS-CoV-2 infection. Fungal and bacterial gut microbiome was characterized by amplicon sequencing on the MiSeq, Illumina's integrated next generation sequencing instrument. A cohort of 201 age- and sex-matched healthy volunteers from the project PRJNA661289 was used as a control group for the bacterial gut microbiota analysis.
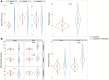
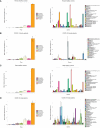
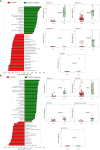
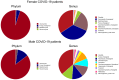
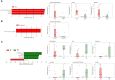
Results: We observed that female COVID-19 patients had a lower gut bacterial microbiota richness than male patients, which was consistent with a different latency in hospital admittance time between the two groups. Both sexes in the COVID-19 patient study group displayed multiple positive associations with opportunistic bacterial pathogens such as Enterococcus, Streptococcus, and Actinomyces. Of note, the Candida genus dominated the gut mycobiota of COVID-19 patients, and adult patients showed a higher intestinal fungal diversity than elderly patients. We found that Saccharomycetales unassigned fungal genera were positively associated with bacterial short-chain fatty acid (SCFA) producers and negatively associated with the proinflammatory genus Bilophila in COVID-19 patients, and we observed that none of the patients who harbored it were admitted to the high-intensity unit.
Conclusions: COVID-19 was associated with opportunistic bacterial pathogens, and Candida was the dominant fungal taxon in the intestine. Together, we found an association between commensal SCFA-producers and a fungal genus that was present in the intestines of patients who did not experience the most severe outcome of the disease. We believe that this taxon could have played a role in the disease outcome, and that further studies should be conducted to understand the role of fungi in gastrointestinal and health protection.
Keywords: COVID-19; SARS-CoV-2; SCFA (short chain fatty acid); human microbiota; microbiome; mycobiota; pathogenesis.
Copyright © 2024 Rizzello, Viciani, Gionchetti, Filippone, Imbesi, Melotti, Dussias, Salice, Santacroce, Padella, Velichevskaya, Marcante and Castagnetti.
Conflict of interest statement
Authors EV, BS, AP, AV, AM, and AC were employed by the company Wellmicro srl. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

