Diverse winter communities and biogeochemical cycling potential in the under-ice microbial plankton of a subarctic river-to-sea continuum
- PMID: 38511950
- PMCID: PMC11210273
- DOI: 10.1128/spectrum.04160-23
Diverse winter communities and biogeochemical cycling potential in the under-ice microbial plankton of a subarctic river-to-sea continuum
Abstract
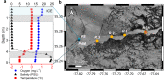
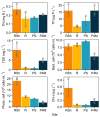
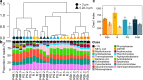
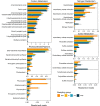
Winter conditions greatly alter the limnological properties of lotic ecosystems and the availability of nutrients, carbon, and energy resources for microbial processes. However, the composition and metabolic capabilities of winter microbial communities are still largely uncharacterized. Here, we sampled the winter under-ice microbiome of the Great Whale River (Nunavik, Canada) and its discharge plume into Hudson Bay. We used a combination of 16S and 18S rRNA gene amplicon analysis and metagenomic sequencing to evaluate the size-fractionated composition and functional potential of the microbial plankton. These under-ice communities were diverse in taxonomic composition and metabolically versatile in terms of energy and carbon acquisition, including the capacity to carry out phototrophic processes and degrade aromatic organic matter. Limnological properties, community composition, and metabolic potential differed between shallow and deeper sites in the river, and between fresh and brackish water in the vertical profile of the plume. Community composition also varied by size fraction, with a greater richness of prokaryotes in the larger size fraction (>3 µm) and of microbial eukaryotes in the smaller size fraction (0.22-3 µm). The freshwater communities included cosmopolitan bacterial genera that were previously detected in the summer, indicating their persistence over time in a wide range of physico-chemical conditions. These observations imply that the microbial communities of subarctic rivers and their associated discharge plumes retain a broad taxonomic and functional diversity throughout the year and that microbial processing of complex terrestrial materials persists beneath the ice during the long winter season.
Importance: Microbiomes vary over multiple timescales, with short- and long-term changes in the physico-chemical environment. However, there is a scarcity of data and understanding about the structure and functioning of aquatic ecosystems during winter relative to summer. This is especially the case for seasonally ice-covered rivers, limiting our understanding of these ecosystems that are common throughout the boreal, subpolar, and polar regions. Here, we examined the winter under-ice microbiome of a Canadian subarctic river and its entry to the sea to characterize the taxonomic and functional features of the microbial community. We found substantial diversity in both composition and functional capabilities, including the capacity to degrade complex terrestrial compounds, despite the constraints imposed by a prolonged seasonal ice-cover and near-freezing water temperatures. This study indicates the ecological complexity and importance of winter microbiomes in ice-covered rivers and the coastal marine environment that they discharge into.
Keywords: coastal water; metagenome; microbial eukaryotes; microbiome; prokaryotes; river; size fraction; subarctic; under-ice; winter limnology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ettema R. 2008. Ice effects on sediments transport in rivers, p 613–648. In Garcia MH (ed), Sedimentation engineering: processes, measurements, modeling, and practice. American Society of Civil Engineers, Virginia.
-
- Bertilsson S, Burgin A, Carey CC, Fey SB, Grossart H-P, Grubisic LM, Jones ID, Kirillin G, Lennon JT, Shade A, Smyth RL. 2013. The under-ice microbiome of seasonally frozen lakes. Limnol Oceanogr 58:1998–2012. doi:10.4319/lo.2013.58.6.1998 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

