A latent clinical-anatomical dimension relating metabolic syndrome to brain structure and cognition
- PMID: 38512127
- PMCID: PMC10957178
- DOI: 10.7554/eLife.93246
A latent clinical-anatomical dimension relating metabolic syndrome to brain structure and cognition
Abstract
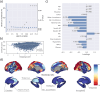
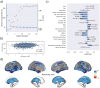
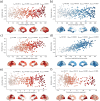
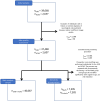
The link between metabolic syndrome (MetS) and neurodegenerative as well as cerebrovascular conditions holds substantial implications for brain health in at-risk populations. This study elucidates the complex relationship between MetS and brain health by conducting a comprehensive examination of cardiometabolic risk factors, brain morphology, and cognitive function in 40,087 individuals. Multivariate, data-driven statistics identified a latent dimension linking more severe MetS to widespread brain morphological abnormalities, accounting for up to 71% of shared variance in the data. This dimension was replicable across sub-samples. In a mediation analysis, we could demonstrate that MetS-related brain morphological abnormalities mediated the link between MetS severity and cognitive performance in multiple domains. Employing imaging transcriptomics and connectomics, our results also suggest that MetS-related morphological abnormalities are linked to the regional cellular composition and macroscopic brain network organization. By leveraging extensive, multi-domain data combined with a dimensional stratification approach, our analysis provides profound insights into the association of MetS and brain health. These findings can inform effective therapeutic and risk mitigation strategies aimed at maintaining brain integrity.
Keywords: brain morphology; cognitive function; connectomics; human; imaging transcriptomics; magnetic resonance imaging; medicine; metabolic syndrome; neuroscience.
© 2023, Petersen et al.
Conflict of interest statement
MP, FH, FN, CM, MS, DR, BZ, SK, AO, KP, SE, BC No competing interests declared, TZ TZ is listed as co-inventor of an international patent on the use of a computing device to estimate the probability of myocardial infarction (PCT/EP2021/073193, International Publication Number WO2022043229A1). TZ is shareholder of the company ART-EMIS GmbH Hamburg, JG JG has received speaker fees from Lundbeck, Janssen-Cilag, Lilly, Otsuka and Boehringer outside the submitted work, JF JF reported receiving personal fees from Acandis, Cerenovus, Microvention, Medtronic, Phenox, and Penumbra; receiving grants from Stryker and Route 92; being managing director of eppdata; and owning shares in Tegus and Vastrax; all outside the submitted work, RT RT is listed as co-inventor of an international patent on the use of a computing device to estimate the probability of myocardial infarction (PCT/EP2021/073193, International Publication Number WO2022043229A1). RT is shareholder of the company ART-EMIS GmbH Hamburg, GT GT has received fees as consultant or lecturer from Acandis, Alexion, Amarin, Bayer, Boehringer Ingelheim, BristolMyersSquibb/Pfizer, Daichi Sankyo, Portola, and Stryker outside the submitted work
Figures
















Update of
-
A latent clinical-anatomical dimension relating metabolic syndrome to brain structure and cognition.bioRxiv [Preprint]. 2023 Dec 23:2023.02.22.529531. doi: 10.1101/2023.02.22.529531. bioRxiv. 2023. Update in: Elife. 2024 Mar 21;12:RP93246. doi: 10.7554/eLife.93246. PMID: 36865285 Free PMC article. Updated. Preprint.
References
-
- Atti AR, Valente S, Iodice A, Caramella I, Ferrari B, Albert U, Mandelli L, De Ronchi D. Metabolic syndrome, mild cognitive impairment, and dementia: A meta-analysis of longitudinal studies. The American Journal of Geriatric Psychiatry. 2019;27:625–637. doi: 10.1016/j.jagp.2019.01.214. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

