The exon junction complex is required for DMD gene splicing fidelity and myogenic differentiation
- PMID: 38512499
- PMCID: PMC10957711
- DOI: 10.1007/s00018-024-05188-1
The exon junction complex is required for DMD gene splicing fidelity and myogenic differentiation
Abstract
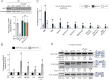
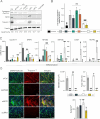
Deposition of the exon junction complex (EJC) upstream of exon-exon junctions helps maintain transcriptome integrity by preventing spurious re-splicing events in already spliced mRNAs. Here we investigate the importance of EJC for the correct splicing of the 2.2-megabase-long human DMD pre-mRNA, which encodes dystrophin, an essential protein involved in cytoskeletal organization and cell signaling. Using targeted RNA-seq, we show that knock-down of the eIF4A3 and Y14 core components of EJC in a human muscle cell line causes an accumulation of mis-splicing events clustered towards the 3' end of the DMD transcript (Dp427m). This deregulation is conserved in the short Dp71 isoform expressed ubiquitously except in adult skeletal muscle and is rescued with wild-type eIF4A3 and Y14 proteins but not with an EJC assembly-defective mutant eIF4A3. MLN51 protein and EJC-associated ASAP/PSAP complexes independently modulate the inclusion of the regulated exons 71 and 78. Our data confirm the protective role of EJC in maintaining splicing fidelity, which in the DMD gene is necessary to preserve the function of the critical C-terminal protein-protein interaction domain of dystrophin present in all tissue-specific isoforms. Given the role of the EJC in maintaining the integrity of dystrophin, we asked whether the EJC could also be involved in the regulation of a mechanism as complex as skeletal muscle differentiation. We found that eIF4A3 knockdown impairs myogenic differentiation by blocking myotube formation. Collectively, our data provide new insights into the functional roles of EJC in human skeletal muscle.
Keywords: Duchenne muscular dystrophy; Myoblasts differentiation; Nonsense mediated mRNA Decay (NMD); PININ; RNPS1; UPF1.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

