Targeted degradation of zDHHC-PATs decreases substrate S-palmitoylation
- PMID: 38512906
- PMCID: PMC10956751
- DOI: 10.1371/journal.pone.0299665
Targeted degradation of zDHHC-PATs decreases substrate S-palmitoylation
Abstract
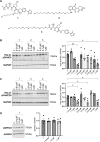
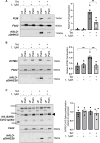
Reversible S-palmitoylation of protein cysteines, catalysed by a family of integral membrane zDHHC-motif containing palmitoyl acyl transferases (zDHHC-PATs), controls the localisation, activity, and interactions of numerous integral and peripheral membrane proteins. There are compelling reasons to want to inhibit the activity of individual zDHHC-PATs in both the laboratory and the clinic, but the specificity of existing tools is poor. Given the extensive conservation of the zDHHC-PAT active site, development of isoform-specific competitive inhibitors is highly challenging. We therefore hypothesised that proteolysis-targeting chimaeras (PROTACs) may offer greater specificity to target this class of enzymes. In proof-of-principle experiments we engineered cell lines expressing tetracycline-inducible Halo-tagged zDHHC5 or zDHHC20, and evaluated the impact of Halo-PROTACs on zDHHC-PAT expression and substrate palmitoylation. In HEK-derived FT-293 cells, Halo-zDHHC5 degradation significantly decreased palmitoylation of its substrate phospholemman, and Halo-zDHHC20 degradation significantly diminished palmitoylation of its substrate IFITM3, but not of the SARS-CoV-2 spike protein. In contrast, in a second kidney derived cell line, Vero E6, Halo-zDHHC20 degradation did not alter palmitoylation of either IFITM3 or SARS-CoV-2 spike. We conclude from these experiments that PROTAC-mediated targeting of zDHHC-PATs to decrease substrate palmitoylation is feasible. However, given the well-established degeneracy in the zDHHC-PAT family, in some settings the activity of non-targeted zDHHC-PATs may substitute and preserve substrate palmitoylation.
Copyright: © 2024 Bai et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
In vitro reconstitution reveals substrate selectivity of protein S-acyltransferases.J Biol Chem. 2025 Apr;301(4):108406. doi: 10.1016/j.jbc.2025.108406. Epub 2025 Mar 14. J Biol Chem. 2025. PMID: 40090582 Free PMC article.
-
The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity.J Biol Chem. 2017 Dec 29;292(52):21517-21526. doi: 10.1074/jbc.M117.800482. Epub 2017 Oct 27. J Biol Chem. 2017. PMID: 29079573 Free PMC article.
-
Spatial organization of palmitoyl acyl transferases governs substrate localization and function.Mol Membr Biol. 2019 Dec;35(1):60-75. doi: 10.1080/09687688.2019.1710274. Mol Membr Biol. 2019. PMID: 31969037 Free PMC article. Review.
-
Control of protein palmitoylation by regulating substrate recruitment to a zDHHC-protein acyltransferase.Commun Biol. 2020 Jul 31;3(1):411. doi: 10.1038/s42003-020-01145-3. Commun Biol. 2020. PMID: 32737405 Free PMC article.
-
Accessory proteins of the zDHHC family of S-acylation enzymes.J Cell Sci. 2020 Nov 17;133(22):jcs251819. doi: 10.1242/jcs.251819. J Cell Sci. 2020. PMID: 33203738 Review.
Cited by
-
Influence of palmitoylation in axonal transport mechanisms in neurodegenerative diseases.Front Cell Neurosci. 2025 Aug 4;19:1613379. doi: 10.3389/fncel.2025.1613379. eCollection 2025. Front Cell Neurosci. 2025. PMID: 40831763 Free PMC article. Review.
-
Recapitulating the potential contribution of protein S-palmitoylation in cancer.Cancer Metastasis Rev. 2024 Dec 27;44(1):20. doi: 10.1007/s10555-024-10217-3. Cancer Metastasis Rev. 2024. PMID: 39725785 Review.
-
Post-translational acylation of proteins in cardiac hypertrophy.Nat Rev Cardiol. 2025 Apr 14. doi: 10.1038/s41569-025-01150-1. Online ahead of print. Nat Rev Cardiol. 2025. PMID: 40229510 Review.
-
Nanobody-thioesterase chimeras to specifically target protein palmitoylation.Nat Commun. 2025 Feb 7;16(1):1445. doi: 10.1038/s41467-025-56716-x. Nat Commun. 2025. PMID: 39920166 Free PMC article.
-
Palmitoylation: an emerging therapeutic target bridging physiology and disease.Cell Mol Biol Lett. 2025 Aug 15;30(1):98. doi: 10.1186/s11658-025-00776-w. Cell Mol Biol Lett. 2025. PMID: 40817227 Free PMC article. Review.
References
-
- Grantham ML, Wu WH, Lalime EN, Lorenzo ME, Klein SL, Pekosz A. Palmitoylation of the influenza A virus M2 protein is not required for virus replication in vitro but contributes to virus virulence. J Virol. 2009;83(17):8655–61. Epub 2009/06/26. doi: 10.1128/JVI.01129-09 ; PubMed Central PMCID: PMC2738213. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

