Adaptive introgression of a visual preference gene
- PMID: 38513020
- PMCID: PMC7616200
- DOI: 10.1126/science.adj9201
Adaptive introgression of a visual preference gene
Abstract
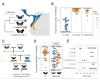
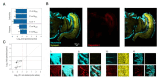
Visual preferences are important drivers of mate choice and sexual selection, but little is known of how they evolve at the genetic level. In this study, we took advantage of the diversity of bright warning patterns displayed by Heliconius butterflies, which are also used during mate choice. Combining behavioral, population genomic, and expression analyses, we show that two Heliconius species have evolved the same preferences for red patterns by exchanging genetic material through hybridization. Neural expression of regucalcin1 correlates with visual preference across populations, and disruption of regucalcin1 with CRISPR-Cas9 impairs courtship toward conspecific females, providing a direct link between gene and behavior. Our results support a role for hybridization during behavioral evolution and show how visually guided behaviors contributing to adaptation and speciation are encoded within the genome.
Conflict of interest statement
Figures


Comment in
-
A genetic cause of male mate preference.Science. 2024 Mar 22;383(6689):1290-1291. doi: 10.1126/science.ado4079. Epub 2024 Mar 21. Science. 2024. PMID: 38513043
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

