Epithelial-derived interleukin-23 promotes oral mucosal immunopathology
- PMID: 38513665
- PMCID: PMC11058479
- DOI: 10.1016/j.immuni.2024.02.020
Epithelial-derived interleukin-23 promotes oral mucosal immunopathology
Abstract
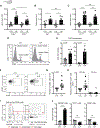
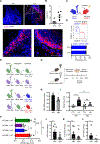
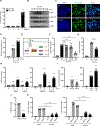
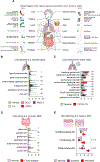
At mucosal surfaces, epithelial cells provide a structural barrier and an immune defense system. However, dysregulated epithelial responses can contribute to disease states. Here, we demonstrated that epithelial cell-intrinsic production of interleukin-23 (IL-23) triggers an inflammatory loop in the prevalent oral disease periodontitis. Epithelial IL-23 expression localized to areas proximal to the disease-associated microbiome and was evident in experimental models and patients with common and genetic forms of disease. Mechanistically, flagellated microbial species of the periodontitis microbiome triggered epithelial IL-23 induction in a TLR5 receptor-dependent manner. Therefore, unlike other Th17-driven diseases, non-hematopoietic-cell-derived IL-23 served as an initiator of pathogenic inflammation in periodontitis. Beyond periodontitis, analysis of publicly available datasets revealed the expression of epithelial IL-23 in settings of infection, malignancy, and autoimmunity, suggesting a broader role for epithelial-intrinsic IL-23 in human disease. Collectively, this work highlights an important role for the barrier epithelium in the induction of IL-23-mediated inflammation.
Keywords: IL-23; Pseudomonas aeruginosa; TLR5; barrier immunity; epithelial; epithelial-intrinsic; flagellin; oral mucosa; pathogenic Th17; periodontitis.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Comment in
-
Gums make IL-23, no professionals needed.Immunity. 2024 Apr 9;57(4):832-834. doi: 10.1016/j.immuni.2024.03.009. Immunity. 2024. PMID: 38599173
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

