PLD1 promotes spindle assembly and migration through regulating autophagy in mouse oocyte meiosis
- PMID: 38513669
- PMCID: PMC11210919
- DOI: 10.1080/15548627.2024.2333164
PLD1 promotes spindle assembly and migration through regulating autophagy in mouse oocyte meiosis
Abstract
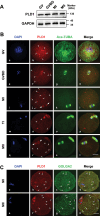
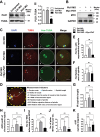
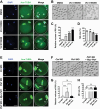
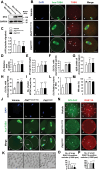
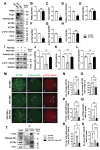
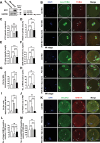
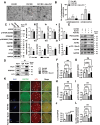
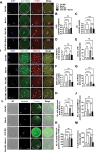
PLD1 has been implicated in cytoskeletal reorganization and vesicle trafficking in somatic cells; however, its function remains unclear in oocyte meiosis. Herein, we found PLD1 stably expresses in mouse oocytes meiosis, with direct interaction with spindle, RAB11A+ vesicles and macroautophagic/autophagic vacuoles. The genetic or chemical inhibition of PLD1 disturbed MTOC clustering, spindle assembly and its cortical migration, also decreased PtdIns(4,5)P2, phosphorylated CFL1 (p-CFL1 [Ser3]) and ACTR2, and their local distribution on MTOC, spindle and vesicles. Furthermore in PLD1-suppressed oocytes, vesicle size was significantly reduced while F-actin density was dramatically increased in the cytoplasm, the asymmetric distribution of autophagic vacuoles was broken and the whole autophagic process was substantially enhanced, as illustrated with characteristic changes in autophagosomes, autolysosome formation and levels of ATG5, BECN1, LC3-II, SQSTM1 and UB. Exogenous administration of PtdIns(4,5)P2 or overexpression of CFL1 hyperphosphorylation mutant (CFL1S3E) could significantly improve polar MTOC focusing and spindle structure in PLD1-depleted oocytes, whereas overexpression of ACTR2 could rescue not only MTOC clustering, and spindle assembly but also its asymmetric positioning. Interestingly, autophagy activation induced similar defects in spindle structure and positioning; instead, its inhibition alleviated the alterations in PLD1-depleted oocytes, and this was highly attributed to the restored levels of PtdIns(4,5)P2, ACTR2 and p-CFL1 (Ser3). Together, PLD1 promotes spindle assembly and migration in oocyte meiosis, by maintaining rational levels of ACTR2, PtdIns(4,5)P2 and p-CFL1 (Ser3) in a manner of modulating autophagy flux. This study for the first time introduces a unique perspective on autophagic activity and function in oocyte meiotic development.Abbreviations: ACTR2/ARP2: actin related protein 2; ACTR3/ARP3: actin related protein 3; ATG5: autophagy related 5; Baf-A1: bafilomycin A1; BFA: brefeldin A; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GOLGA2/GM130: golgin A2; GV: germinal vesicle; GVBD: germinal vesicle breakdown; IVM: in vitro maturation; MAP1LC3/LC3: microtubule-associated protein 1 light chain 3; MI: metaphase of meiosis I; MII: metaphase of meiosis II; MO: morpholino; MTOC: microtubule-organizing center; MTOR: mechanistic target of rapamycin kinase; PB1: first polar body; PLA: proximity ligation assay; PLD1: phospholipase D1; PtdIns(4,5)P2/PIP2: phosphatidylinositol 4,5-bisphosphate; RAB11A: RAB11A, member RAS oncogene family; RPS6KB1/S6K1: ribosomal protein S6 kinase B1; SQSTM1/p62: sequestosome 1; TEM: transmission electron microscopy; TUBA/α-tubulin: tubulin alpha; TUBG/γ-tubulin: tubulin gamma; UB: ubiquitin; WASL/N-WASP: WASP like actin nucleation promoting factor.
Keywords: Autophagy; MTOC clustering; PLD1; meiosis; oocyte; spindle migration.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Coticchio G, Dal Canto M, Mignini Renzini M, et al. Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum Reprod Update. 2015. Jul-Aug;21(4):427–454. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
