Gasdermin B, an asthma-susceptibility gene, promotes MAVS-TBK1 signalling and airway inflammation
- PMID: 38514093
- PMCID: PMC11063620
- DOI: 10.1183/13993003.01232-2023
Gasdermin B, an asthma-susceptibility gene, promotes MAVS-TBK1 signalling and airway inflammation
Abstract
Rationale: Respiratory virus-induced inflammation is the leading cause of asthma exacerbation, frequently accompanied by induction of interferon-stimulated genes (ISGs). How asthma-susceptibility genes modulate cellular response upon viral infection by fine-tuning ISG induction and subsequent airway inflammation in genetically susceptible asthma patients remains largely unknown.
Objectives: To decipher the functions of gasdermin B (encoded by GSDMB) in respiratory virus-induced lung inflammation.
Methods: In two independent cohorts, we analysed expression correlation between GSDMB and ISG s. In human bronchial epithelial cell line or primary bronchial epithelial cells, we generated GSDMB-overexpressing and GSDMB-deficient cells. A series of quantitative PCR, ELISA and co-immunoprecipitation assays were performed to determine the function and mechanism of GSDMB for ISG induction. We also generated a novel transgenic mouse line with inducible expression of human unique GSDMB gene in airway epithelial cells and infected the mice with respiratory syncytial virus to determine the role of GSDMB in respiratory syncytial virus-induced lung inflammation in vivo.
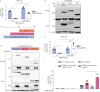
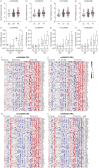
Results: GSDMB is one of the most significant asthma-susceptibility genes at 17q21 and acts as a novel RNA sensor, promoting mitochondrial antiviral-signalling protein (MAVS)-TANK binding kinase 1 (TBK1) signalling and subsequent inflammation. In airway epithelium, GSDMB is induced by respiratory viral infections. Expression of GSDMB and ISGs significantly correlated in respiratory epithelium from two independent asthma cohorts. Notably, inducible expression of human GSDMB in mouse airway epithelium led to enhanced ISGs induction and increased airway inflammation with mucus hypersecretion upon respiratory syncytial virus infection.
Conclusions: GSDMB promotes ISGs expression and airway inflammation upon respiratory virus infection, thereby conferring asthma risk in risk allele carriers.
Copyright ©The authors 2024.
Conflict of interest statement
Conflict of interest: The authors have no potential conflicts of interest to disclose.
Figures





Comment in
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
