Dilation of ion selectivity filters in cation channels
- PMID: 38514273
- PMCID: PMC11069442
- DOI: 10.1016/j.tibs.2024.02.004
Dilation of ion selectivity filters in cation channels
Abstract
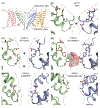
Ion channels establish the voltage gradient across cellular membranes by providing aqueous pathways for ions to selectively diffuse down their concentration gradients. The selectivity of any given channel for its favored ions has conventionally been viewed as a stable property, and in many cation channels, it is determined by an ion-selectivity filter within the external end of the ion-permeation pathway. In several instances, including voltage-activated K+ (Kv) channels, ATP-activated P2X receptor channels, and transient receptor potential (TRP) channels, the ion-permeation pathways have been proposed to dilate in response to persistent activation, dynamically altering ion permeation. Here, we discuss evidence for dynamic ion selectivity, examples where ion selectivity filters exhibit structural plasticity, and opportunities to fill gaps in our current understanding.
Keywords: ATP-activated P2X receptor channels; Kv channels; TRP channels; ion selectivity.
Published by Elsevier Ltd.
Conflict of interest statement
Declaration of interests No interests are declared.
Figures
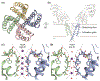
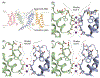

References
-
- Hille B. (2001) Ion channels of excitable membranes 3rd edn), Sinauer
-
- MacKinnon R and Miller C (1989) Mutant potassium channels with altered binding of charybdotoxin, a pore- blocking peptide inhibitor. Science 245, 1382–1385 - PubMed
-
- Yellen G. et al. (1991) Mutations affecting internal TEA blockade identify the probable pore- forming region of a K+ channel. Science 251, 939–942 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

