Advancing entity recognition in biomedicine via instruction tuning of large language models
- PMID: 38514400
- PMCID: PMC11001490
- DOI: 10.1093/bioinformatics/btae163
Advancing entity recognition in biomedicine via instruction tuning of large language models
Abstract
Motivation: Large Language Models (LLMs) have the potential to revolutionize the field of Natural Language Processing, excelling not only in text generation and reasoning tasks but also in their ability for zero/few-shot learning, swiftly adapting to new tasks with minimal fine-tuning. LLMs have also demonstrated great promise in biomedical and healthcare applications. However, when it comes to Named Entity Recognition (NER), particularly within the biomedical domain, LLMs fall short of the effectiveness exhibited by fine-tuned domain-specific models. One key reason is that NER is typically conceptualized as a sequence labeling task, whereas LLMs are optimized for text generation and reasoning tasks.
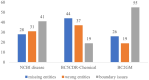
Results: We developed an instruction-based learning paradigm that transforms biomedical NER from a sequence labeling task into a generation task. This paradigm is end-to-end and streamlines the training and evaluation process by automatically repurposing pre-existing biomedical NER datasets. We further developed BioNER-LLaMA using the proposed paradigm with LLaMA-7B as the foundational LLM. We conducted extensive testing on BioNER-LLaMA across three widely recognized biomedical NER datasets, consisting of entities related to diseases, chemicals, and genes. The results revealed that BioNER-LLaMA consistently achieved higher F1-scores ranging from 5% to 30% compared to the few-shot learning capabilities of GPT-4 on datasets with different biomedical entities. We show that a general-domain LLM can match the performance of rigorously fine-tuned PubMedBERT models and PMC-LLaMA, biomedical-specific language model. Our findings underscore the potential of our proposed paradigm in developing general-domain LLMs that can rival SOTA performances in multi-task, multi-domain scenarios in biomedical and health applications.
Availability and implementation: Datasets and other resources are available at https://github.com/BIDS-Xu-Lab/BioNER-LLaMA.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
The authors do not have any conflicts of interest to disclose.
Figures



Similar articles
-
Resource-efficient instruction tuning of large language models for biomedical named entity recognition.J Biomed Inform. 2025 Aug 21;170:104896. doi: 10.1016/j.jbi.2025.104896. Online ahead of print. J Biomed Inform. 2025. PMID: 40849052
-
BioInstruct: instruction tuning of large language models for biomedical natural language processing.J Am Med Inform Assoc. 2024 Sep 1;31(9):1821-1832. doi: 10.1093/jamia/ocae122. J Am Med Inform Assoc. 2024. PMID: 38833265 Free PMC article.
-
Me-LLaMA: Foundation Large Language Models for Medical Applications.Res Sq [Preprint]. 2024 May 22:rs.3.rs-4240043. doi: 10.21203/rs.3.rs-4240043/v1. Res Sq. 2024. PMID: 38826372 Free PMC article. Preprint.
-
Applications and Concerns of ChatGPT and Other Conversational Large Language Models in Health Care: Systematic Review.J Med Internet Res. 2024 Nov 7;26:e22769. doi: 10.2196/22769. J Med Internet Res. 2024. PMID: 39509695 Free PMC article.
-
Examining the Role of Large Language Models in Orthopedics: Systematic Review.J Med Internet Res. 2024 Nov 15;26:e59607. doi: 10.2196/59607. J Med Internet Res. 2024. PMID: 39546795 Free PMC article.
Cited by
-
HunFlair2 in a cross-corpus evaluation of biomedical named entity recognition and normalization tools.Bioinformatics. 2024 Oct 1;40(10):btae564. doi: 10.1093/bioinformatics/btae564. Bioinformatics. 2024. PMID: 39302686 Free PMC article.
-
Evaluation of SURUS: a named entity recognition NLP system to extract knowledge from interventional study records.BMC Med Res Methodol. 2025 Jul 31;25(1):184. doi: 10.1186/s12874-025-02624-z. BMC Med Res Methodol. 2025. PMID: 40745274 Free PMC article.
-
A foundation model for human-AI collaboration in medical literature mining.ArXiv [Preprint]. 2025 Jan 27:arXiv:2501.16255v1. ArXiv. 2025. PMID: 40735107 Free PMC article. Preprint.
-
Benchmarking large language models for biomedical natural language processing applications and recommendations.Nat Commun. 2025 Apr 6;16(1):3280. doi: 10.1038/s41467-025-56989-2. Nat Commun. 2025. PMID: 40188094 Free PMC article.
-
Toward Cross-Hospital Deployment of Natural Language Processing Systems: Model Development and Validation of Fine-Tuned Large Language Models for Disease Name Recognition in Japanese.JMIR Med Inform. 2025 Jul 8;13:e76773. doi: 10.2196/76773. JMIR Med Inform. 2025. PMID: 40627819 Free PMC article.
References
-
- Achiam J, Adler S, Agarwal S. et al. Gpt-4 technical report. arXiv, arXiv:2303.08774, 2023, preprint: not peer reviewed.
-
- Agrawal M, Hegselmann S, Lang H. et al. Large language models are few-shot clinical information extractors. In: Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing, Abu Dhabi, UAE. 2022.
-
- Ashok D, Lipton ZC. PromptNER: prompting for named entity recognition. arXiv, arXiv:2305.15444, 2023, preprint: not peer reviewed.
-
- Beltagy I, Lo K, Cohan A. SciBERT: a pretrained language model for scientific text. In: Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), pp. 3615–3620, Hong Kong, China. Association for Computational Linguistics. 2019.
-
- Biderman S, Schoelkopf H, Anthony QG. et al. Pythia: a suite for analyzing large language models across training and scaling. In: International Conference on Machine Learning, Honolulu, Hawaii, USA. 2023.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

