Dual RNA sequencing of Helicobacter pylori and host cell transcriptomes reveals ontologically distinct host-pathogen interaction
- PMID: 38514462
- PMCID: PMC11019886
- DOI: 10.1128/msystems.00206-24
Dual RNA sequencing of Helicobacter pylori and host cell transcriptomes reveals ontologically distinct host-pathogen interaction
Abstract
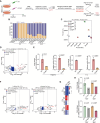
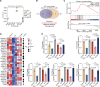
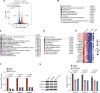
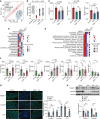
Helicobacter pylori is a highly successful pathogen that poses a substantial threat to human health. However, the dynamic interaction between H. pylori and the human gastric epithelium has not been fully investigated. In this study, using dual RNA sequencing technology, we characterized a cytotoxin-associated gene A (cagA)-modulated bacterial adaption strategy by enhancing the expression of ATP-binding cassette transporter-related genes, metQ and HP_0888, upon coculturing with human gastric epithelial cells. We observed a general repression of electron transport-associated genes by cagA, leading to the activation of oxidative phosphorylation. Temporal profiling of host mRNA signatures revealed the downregulation of multiple splicing regulators due to bacterial infection, resulting in aberrant pre-mRNA splicing of functional genes involved in the cell cycle process in response to H. pylori infection. Moreover, we demonstrated a protective effect of gastric H. pylori colonization against chronic dextran sulfate sodium (DSS)-induced colitis. Mechanistically, we identified a cluster of propionic and butyric acid-producing bacteria, Muribaculaceae, selectively enriched in the colons of H. pylori-pre-colonized mice, which may contribute to the restoration of intestinal barrier function damaged by DSS treatment. Collectively, this study presents the first dual-transcriptome analysis of H. pylori during its dynamic interaction with gastric epithelial cells and provides new insights into strategies through which H. pylori promotes infection and pathogenesis in the human gastric epithelium.
Importance: Simultaneous profiling of the dynamic interaction between Helicobacter pylori and the human gastric epithelium represents a novel strategy for identifying regulatory responses that drive pathogenesis. This study presents the first dual-transcriptome analysis of H. pylori when cocultured with gastric epithelial cells, revealing a bacterial adaptation strategy and a general repression of electron transportation-associated genes, both of which were modulated by cytotoxin-associated gene A (cagA). Temporal profiling of host mRNA signatures dissected the aberrant pre-mRNA splicing of functional genes involved in the cell cycle process in response to H. pylori infection. We demonstrated a protective effect of gastric H. pylori colonization against chronic DSS-induced colitis through both in vitro and in vivo experiments. These findings significantly enhance our understanding of how H. pylori promotes infection and pathogenesis in the human gastric epithelium and provide evidence to identify targets for antimicrobial therapies.
Keywords: ATP-binding cassette transporter; Helicobacter pylori; alternative splicing; cytotoxin-associated genes A; dual RNA sequencing; inflammatory bowel disease; oxidative phosphorylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The Helicobacter pylori cytotoxin CagA is essential for suppressing host heat shock protein expression.Cell Stress Chaperones. 2016 May;21(3):523-33. doi: 10.1007/s12192-016-0680-x. Epub 2016 Mar 1. Cell Stress Chaperones. 2016. PMID: 26928021 Free PMC article.
-
The HopQ-CEACAM Interaction Controls CagA Translocation, Phosphorylation, and Phagocytosis of Helicobacter pylori in Neutrophils.mBio. 2020 Feb 4;11(1):e03256-19. doi: 10.1128/mBio.03256-19. mBio. 2020. PMID: 32019805 Free PMC article.
-
CagA-ASPP2 complex mediates loss of cell polarity and favors H. pylori colonization of human gastric organoids.Proc Natl Acad Sci U S A. 2020 Feb 4;117(5):2645-2655. doi: 10.1073/pnas.1908787117. Epub 2020 Jan 21. Proc Natl Acad Sci U S A. 2020. PMID: 31964836 Free PMC article.
-
[Advances in CagA protein and CagAmediated pathogenesis of Helicobacter pylori-A review].Wei Sheng Wu Xue Bao. 2016 Dec 4;56(12):1821-30. Wei Sheng Wu Xue Bao. 2016. PMID: 29741846 Review. Chinese.
-
Influence of Helicobacter pylori virulence factors CagA and VacA on pathogenesis of gastrointestinal disorders.Microb Pathog. 2018 Apr;117:43-48. doi: 10.1016/j.micpath.2018.02.016. Epub 2018 Feb 9. Microb Pathog. 2018. PMID: 29432909 Review.
Cited by
-
The role of BUD31 in clear cell renal cell carcinoma: prognostic significance, alternative splicing, and tumor immune environment.Clin Exp Med. 2024 Aug 13;24(1):191. doi: 10.1007/s10238-024-01451-8. Clin Exp Med. 2024. PMID: 39136845 Free PMC article.
-
Debate on the relationship between Helicobacter pylori infection and inflammatory bowel disease: a bibliometric analysis.Front Microbiol. 2024 Nov 6;15:1479941. doi: 10.3389/fmicb.2024.1479941. eCollection 2024. Front Microbiol. 2024. PMID: 39569001 Free PMC article.
References
-
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi:10.1016/S1473-3099(17)30753-3 - DOI - PubMed
-
- Correa P. 1988. A human model of gastric carcinogenesis. Cancer Res 48:3554–3560. - PubMed
MeSH terms
Substances
Grants and funding
- 81974070/MOST | National Natural Science Foundation of China (NSFC)
- 2020A1515011063, 2023A1515011685, 2023A1515030071/Guangdong basic and applied basic research foundation
- 2020KQNCX013/Guangdong young innovative talents foundation
- JCYJ20210324131010027, JCYJ20220530154205011, JCYJ20230807142314030/Shenzhen Municipal Science and Technology Innovation Council | Shenzhen Science and Technology Innovation Program ()
- PT2018GZR05, PT2018GZR10/Research Foundation of Shenzhen Hospital of Southern Medical University
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

