The EIF3H-HAX1 axis increases RAF-MEK-ERK signaling activity to promote colorectal cancer progression
- PMID: 38514606
- PMCID: PMC10957977
- DOI: 10.1038/s41467-024-46521-3
The EIF3H-HAX1 axis increases RAF-MEK-ERK signaling activity to promote colorectal cancer progression
Abstract
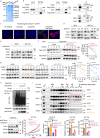
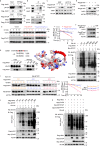
Eukaryotic initiation translation factor 3 subunit h (EIF3H) plays critical roles in regulating translational initiation and predicts poor cancer prognosis, but the mechanism underlying EIF3H tumorigenesis remains to be further elucidated. Here, we report that EIF3H is overexpressed in colorectal cancer (CRC) and correlates with poor prognosis. Conditional Eif3h deletion suppresses colorectal tumorigenesis in AOM/DSS model. Mechanistically, EIF3H functions as a deubiquitinase for HAX1 and stabilizes HAX1 via antagonizing βTrCP-mediated ubiquitination, which enhances the interaction between RAF1, MEK1 and ERK1, thereby potentiating phosphorylation of ERK1/2. In addition, activation of Wnt/β-catenin signaling induces EIF3H expression. EIF3H/HAX1 axis promotes CRC tumorigenesis and metastasis in mouse orthotopic cancer model. Significantly, combined targeting Wnt and RAF1-ERK1/2 signaling synergistically inhibits tumor growth in EIF3H-high patient-derived xenografts. These results uncover the important roles of EIF3H in mediating CRC progression through regulating HAX1 and RAF1-ERK1/2 signaling. EIF3H represents a promising therapeutic target and prognostic marker in CRC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

