Tertiary lymphoid structures and B cells determine clinically relevant T cell phenotypes in ovarian cancer
- PMID: 38514660
- PMCID: PMC10957872
- DOI: 10.1038/s41467-024-46873-w
Tertiary lymphoid structures and B cells determine clinically relevant T cell phenotypes in ovarian cancer
Abstract
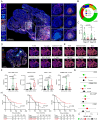
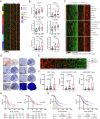
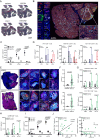
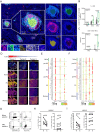
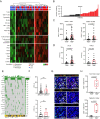
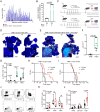
Intratumoral tertiary lymphoid structures (TLSs) have been associated with improved outcome in various cohorts of patients with cancer, reflecting their contribution to the development of tumor-targeting immunity. Here, we demonstrate that high-grade serous ovarian carcinoma (HGSOC) contains distinct immune aggregates with varying degrees of organization and maturation. Specifically, mature TLSs (mTLS) as forming only in 16% of HGSOCs with relatively elevated tumor mutational burden (TMB) are associated with an increased intratumoral density of CD8+ effector T (TEFF) cells and TIM3+PD1+, hence poorly immune checkpoint inhibitor (ICI)-sensitive, CD8+ T cells. Conversely, CD8+ T cells from immunologically hot tumors like non-small cell lung carcinoma (NSCLC) are enriched in ICI-responsive TCF1+ PD1+ T cells. Spatial B-cell profiling identifies patterns of in situ maturation and differentiation associated with mTLSs. Moreover, B-cell depletion promotes signs of a dysfunctional CD8+ T cell compartment among tumor-infiltrating lymphocytes from freshly isolated HGSOC and NSCLC biopsies. Taken together, our data demonstrate that - at odds with NSCLC - HGSOC is associated with a low density of follicular helper T cells and thus develops a limited number of mTLS that might be insufficient to preserve a ICI-sensitive TCF1+PD1+ CD8+ T cell phenotype. These findings point to key quantitative and qualitative differences between mTLSs in ICI-responsive vs ICI-irresponsive neoplasms that may guide the development of alternative immunotherapies for patients with HGSOC.
© 2024. The Author(s).
Conflict of interest statement
L.G. is/has been holding research contracts with Lytix Biopharma, Onxeo and Promontory, has received consulting/advisory honoraria from Boehringer Ingelheim, AstraZeneca, OmniSEQ, Onxeo, The Longevity Labs, Inzen, Sotio, Promontory, Noxopharm and the Luke Heller TECPR2 Foundation, and holds Promontory stock options. R.Sp. is minority shareholder of Sotio Biotech a.s. A.R. declare advisory services and invited lectures for Amgen, AstraZeneca, BMS, Eli-Lilly, Janssen-Cilag, MSD, Roche. All other authors have no conflicts to declare.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

