Focal adhesions contain three specialized actin nanoscale layers
- PMID: 38514695
- PMCID: PMC10957975
- DOI: 10.1038/s41467-024-46868-7
Focal adhesions contain three specialized actin nanoscale layers
Abstract
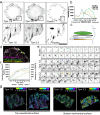
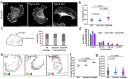
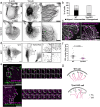
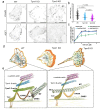
Focal adhesions (FAs) connect inner workings of cell to the extracellular matrix to control cell adhesion, migration and mechanosensing. Previous studies demonstrated that FAs contain three vertical layers, which connect extracellular matrix to the cytoskeleton. By using super-resolution iPALM microscopy, we identify two additional nanoscale layers within FAs, specified by actin filaments bound to tropomyosin isoforms Tpm1.6 and Tpm3.2. The Tpm1.6-actin filaments, beneath the previously identified α-actinin cross-linked actin filaments, appear critical for adhesion maturation and controlled cell motility, whereas the adjacent Tpm3.2-actin filament layer beneath seems to facilitate adhesion disassembly. Mechanistically, Tpm3.2 stabilizes ACF-7/MACF1 and KANK-family proteins at adhesions, and hence targets microtubule plus-ends to FAs to catalyse their disassembly. Tpm3.2 depletion leads to disorganized microtubule network, abnormally stable FAs, and defects in tail retraction during migration. Thus, FAs are composed of distinct actin filament layers, and each may have specific roles in coupling adhesions to the cytoskeleton, or in controlling adhesion dynamics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

