The growth factor EPIREGULIN promotes basal progenitor cell proliferation in the developing neocortex
- PMID: 38514807
- PMCID: PMC11021537
- DOI: 10.1038/s44318-024-00068-7
The growth factor EPIREGULIN promotes basal progenitor cell proliferation in the developing neocortex
Abstract
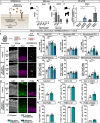
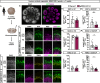
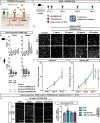
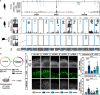
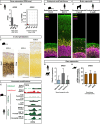
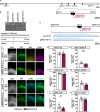
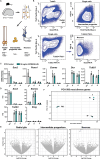
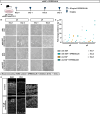
Neocortex expansion during evolution is linked to higher numbers of neurons, which are thought to result from increased proliferative capacity and neurogenic potential of basal progenitor cells during development. Here, we show that EREG, encoding the growth factor EPIREGULIN, is expressed in the human developing neocortex and in gorilla cerebral organoids, but not in the mouse neocortex. Addition of EPIREGULIN to the mouse neocortex increases proliferation of basal progenitor cells, whereas EREG ablation in human cortical organoids reduces proliferation in the subventricular zone. Treatment of cortical organoids with EPIREGULIN promotes a further increase in proliferation of gorilla but not of human basal progenitor cells. EPIREGULIN competes with the epidermal growth factor (EGF) to promote proliferation, and inhibition of the EGF receptor abrogates the EPIREGULIN-mediated increase in basal progenitor cells. Finally, we identify putative cis-regulatory elements that may contribute to the observed inter-species differences in EREG expression. Our findings suggest that species-specific regulation of EPIREGULIN expression may contribute to the increased neocortex size of primates by providing a tunable pro-proliferative signal to basal progenitor cells in the subventricular zone.
Keywords: Cortical Organoid; Gene Regulation; Human Neurogenesis; Neocortex Evolution; Neural Progenitor Cell.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

