Identification of FasL as a crucial host factor driving COVID-19 pathology and lethality
- PMID: 38514848
- PMCID: PMC11093991
- DOI: 10.1038/s41418-024-01278-6
Identification of FasL as a crucial host factor driving COVID-19 pathology and lethality
Abstract
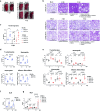
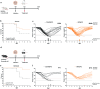
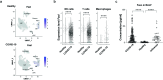
The dysregulated immune response and inflammation resulting in severe COVID-19 are still incompletely understood. Having recently determined that aberrant death-ligand-induced cell death can cause lethal inflammation, we hypothesized that this process might also cause or contribute to inflammatory disease and lung failure following SARS-CoV-2 infection. To test this hypothesis, we developed a novel mouse-adapted SARS-CoV-2 model (MA20) that recapitulates key pathological features of COVID-19. Concomitantly with occurrence of cell death and inflammation, FasL expression was significantly increased on inflammatory monocytic macrophages and NK cells in the lungs of MA20-infected mice. Importantly, therapeutic FasL inhibition markedly increased survival of both, young and old MA20-infected mice coincident with substantially reduced cell death and inflammation in their lungs. Intriguingly, FasL was also increased in the bronchoalveolar lavage fluid of critically-ill COVID-19 patients. Together, these results identify FasL as a crucial host factor driving the immuno-pathology that underlies COVID-19 severity and lethality, and imply that patients with severe COVID-19 may significantly benefit from therapeutic inhibition of FasL.
© 2024. The Author(s).
Conflict of interest statement
HW is co-founder of Apogenix, a biotech company that develops asunercept. The authors declare no other competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- SFB1399, Project C06/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 214342/WT_/Wellcome Trust/United Kingdom
- PID2019-105451GB-I00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- PID2020-113963RBI00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- SFB1403-414786233/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

