STING is significantly increased in high-grade glioma with high risk of recurrence
- PMID: 38516268
- PMCID: PMC10956623
- DOI: 10.1080/2162402X.2024.2327682
STING is significantly increased in high-grade glioma with high risk of recurrence
Abstract
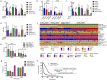
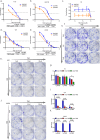
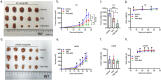
In this study, we aimed to comprehensively characterize the potential relationships among the frequently mutated genes, well-known homologous recombination repair (HRR) proteins, and immune proteins in glioma from a clinical perspective. A total of 126 surgical tissues from patients initially diagnosed with glioma were included. The genetic alterations were tested using the targeted next-generation sequencing technique. The expression of HRR proteins, immune proteins, and genetic alteration-related proteins were detected using immunostaining. Integrated analysis showed that ATRX is positively correlated with STING in high-grade glioma (HGG) with wild-type ATRX and IDH1. Then, a relapse predictive risk-scoring model was established using the least absolute shrinkage and selection operator regression algorithms. The scores based on the expression of ATRX and STING significantly predict the recurrence for glioma patients, which further predict the survival for specific subgroups, characterized with high expression of RAD51 and wild-type TERT. Moreover, STING is significantly higher in patients with high relapse risk. Interestingly, STING inhibitors and agonists both suppress the growth of HGG cells, regardless of their STING levels and STING pathway activity, whereas RAD51 inhibitor B02 is found to exclusively sensitize HGG cells with high expression of STING to temozolomide in vitro and in vivo. Overall, findings in the study not only reveal that ATRX is closely correlated with STING to drive the relapse of HGG, but also provide a STING-guided combined strategy to treat patients with aggressive gliomas. Translation of these findings will ultimately improve the outcomes for ATRX and IDH1 genomically stratified subgroups in HGG.
Keywords: ATRX; IDH1; RAD51 inhibitor; STING; high-grade glioma; temozolomide sensitivity.
© 2024 The Author(s). Published with license by Taylor & Francis Group, LLC.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
Main genetic differences in high-grade gliomas may present different MR imaging and MR spectroscopy correlates.Eur Radiol. 2021 Feb;31(2):749-763. doi: 10.1007/s00330-020-07138-4. Epub 2020 Sep 1. Eur Radiol. 2021. PMID: 32875375
-
Detection of ATRX and IDH1-R132H immunohistochemistry in the progression of 211 paired gliomas.Oncotarget. 2016 Mar 29;7(13):16384-95. doi: 10.18632/oncotarget.7650. Oncotarget. 2016. PMID: 26918938 Free PMC article.
-
Mutant IDH1 Cooperates with ATRX Loss to Drive the Alternative Lengthening of Telomere Phenotype in Glioma.Cancer Res. 2018 Jun 1;78(11):2966-2977. doi: 10.1158/0008-5472.CAN-17-2269. Epub 2018 Mar 15. Cancer Res. 2018. PMID: 29545335 Free PMC article.
-
New Molecular Considerations for Glioma: IDH, ATRX, BRAF, TERT, H3 K27M.Curr Neurol Neurosci Rep. 2017 Feb;17(2):19. doi: 10.1007/s11910-017-0722-5. Curr Neurol Neurosci Rep. 2017. PMID: 28271343 Review.
-
Molecular genetics of low-grade gliomas: genomic alterations guiding diagnosis and therapeutic intervention. 11th annual Frye-Halloran Brain Tumor Symposium.Neurosurg Focus. 2013 Feb;34(2):E9. doi: 10.3171/2012.12.FOCUS12349. Neurosurg Focus. 2013. PMID: 23373454 Review.
Cited by
-
The STING Signaling: A Novel Target for Central Nervous System Diseases.Cell Mol Neurobiol. 2025 Apr 7;45(1):33. doi: 10.1007/s10571-025-01550-4. Cell Mol Neurobiol. 2025. PMID: 40195137 Free PMC article. Review.
-
KDM1A epigenetically enhances RAD51 expression to suppress the STING-associated anti-tumor immunity in esophageal squamous cell carcinoma.Cell Death Dis. 2024 Dec 6;15(12):882. doi: 10.1038/s41419-024-07275-4. Cell Death Dis. 2024. PMID: 39638799 Free PMC article.
References
-
- Haase S, Banerjee K, Mujeeb AA, Hartlage CS, Núñez FM, Núñez FJ, Alghamri MS, Kadiyala P, Carney S, Barissi MN. et al. H3.3-G34 mutations impair DNA repair and promote cGAS/STING-mediated immune responses in pediatric high-grade glioma models. J Clin Invest. Nov 15 2022;132(22). doi:10.1172/JCI154229. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
