ctDNA transiting into urine is ultrashort and facilitates noninvasive liquid biopsy of HPV+ oropharyngeal cancer
- PMID: 38516891
- PMCID: PMC11018327
- DOI: 10.1172/jci.insight.177759
ctDNA transiting into urine is ultrashort and facilitates noninvasive liquid biopsy of HPV+ oropharyngeal cancer
Abstract
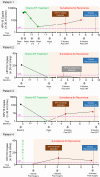
BACKGROUNDTransrenal cell-free tumor DNA (TR-ctDNA), which transits from the bloodstream into urine, has the potential to enable noninvasive cancer detection for a wide variety of nonurologic cancer types.MethodsUsing whole-genome sequencing, we discovered that urine TR-ctDNA fragments across multiple cancer types are predominantly ultrashort (<50 bp) and, therefore, likely to be missed by conventional ctDNA assays. We developed an ultrashort droplet digital PCR assay to detect TR-ctDNA originating from HPV-associated oropharyngeal squamous cell carcinoma (HPV+ OPSCC) and confirmed that assaying ultrashort DNA is critical for sensitive cancer detection from urine samples.ResultsTR-ctDNA was concordant with plasma ctDNA for cancer detection in patients with HPV+ OPSCC. As proof of concept for using urine TR-ctDNA for posttreatment surveillance, in a small longitudinal case series, TR-ctDNA showed promise for noninvasive detection of recurrence of HPV+ OPSCC.ConclusionOur data indicate that focusing on ultrashort fragments of TR-ctDNA will be important for realizing the full potential of urine-based cancer diagnostics. This has implications for urine-based detection of a wide variety of cancer types and for facilitating access to care through at-home specimen collections.FundingNIH grants R33 CA229023, R21 CA225493; NIH/National Cancer Institute grants U01 CA183848, R01 CA184153, and P30CA046592; American Cancer Society RSG-18-062-01-TBG; American Cancer Society Mission Boost grant MBGI-22-056-01-MBG; and the A. Alfred Taubman Medical Research Institute.
Keywords: Cancer; Diagnostics; Head and neck cancer; Oncology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

