Determination of minimum inhibitory concentrations using machine-learning-assisted agar dilution
- PMID: 38517194
- PMCID: PMC11064640
- DOI: 10.1128/spectrum.04209-23
Determination of minimum inhibitory concentrations using machine-learning-assisted agar dilution
Abstract
Effective policy to address the global threat of antimicrobial resistance requires robust antimicrobial susceptibility data. Traditional methods for measuring minimum inhibitory concentration (MIC) are resource intensive, subject to human error, and require considerable infrastructure. AIgarMIC streamlines and standardizes MIC measurement and is especially valuable for large-scale surveillance activities. MICs were measured using agar dilution for n = 10 antibiotics against clinical Enterobacterales isolates (n = 1,086) obtained from a large tertiary hospital microbiology laboratory. Escherichia coli (n = 827, 76%) was the most common organism. Photographs of agar plates were divided into smaller images covering one inoculation site. A labeled data set of colony images was created and used to train a convolutional neural network to classify images based on whether a bacterial colony was present (first-step model). If growth was present, a second-step model determined whether colony morphology suggested antimicrobial growth inhibition. The ability of the AI to determine MIC was then compared with standard visual determination. The first-step model classified bacterial growth as present/absent with 94.3% accuracy. The second-step model classified colonies as "inhibited" or "good growth" with 88.6% accuracy. For the determination of MIC, the rate of essential agreement was 98.9% (644/651), with a bias of -7.8%, compared with manual annotation. AIgarMIC uses artificial intelligence to automate endpoint assessments for agar dilution and potentially increases throughput without bespoke equipment. AIgarMIC reduces laboratory barriers to generating high-quality MIC data that can be used for large-scale surveillance programs.
Importance: This research uses modern artificial intelligence and machine-learning approaches to standardize and automate the interpretation of agar dilution minimum inhibitory concentration testing. Artificial intelligence is currently of significant topical interest to researchers and clinicians. In our manuscript, we demonstrate a use-case in the microbiology laboratory and present validation data for the model's performance against manual interpretation.
Keywords: antimicrobial resistance; artificial intelligence; assay validation; digital health; image recognition; laboratory software; machine learning; minimum inhibitory concentration.
Conflict of interest statement
A.G. holds a research grant with United Kingdom Research and Innovation (UKRI) for technology described in this manuscript. W.H. and A.G. hold research grants with EPSRC and MRC for technology described in this manuscript. A.H. declares consulting work for Pfizer outside of the submitted work. W.H. holds or has held research grants with UKRI, EU, F2G, Spero Therapeutics, Antabio, Pfizer, Bugworks, Phico Therapeutics, BioVersys, Global Antibiotic Research & Development Partnership (GARDP), and NAEJA- RGM. W.H. is or has been a consultant for Appili Therapeutics, F2G, Spero Therapeutics, NAEJA-RGM, Centauri, Pfizer, Phico Therapeutics, Pulmocide, Amplyx, Mundipharma Research, and VenatoRx. W.H. is a member of the Specialist Advisory Committee for GARDP and the Specialty National co-lead for Infectious Diseases for the National Institute of Health Research. All other authors declare no competing interests.
Figures

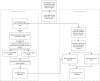




References
-
- CLSI . 2023. Performance standards for antimicrobial susceptibility testing. In CLSI supplement M100, 33rd ed. CLSI supplement M100.
-
- The European Committee on Antimicrobial Susceptibility Testing . 2023. EUCAST Breakpoint tables (13.1)
-
- Pfizer . Antimicrobial testing leadership and surveillance (ATLAS). Available from: https://atlas-surveillance.com. Retrieved 30 Aug 2023.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

