Deep learning-based 3D cerebrovascular segmentation workflow on bright and black blood sequences magnetic resonance angiography
- PMID: 38517610
- PMCID: PMC10959883
- DOI: 10.1186/s13244-024-01657-0
Deep learning-based 3D cerebrovascular segmentation workflow on bright and black blood sequences magnetic resonance angiography
Abstract
Background: Cerebrovascular diseases have emerged as significant threats to human life and health. Effectively segmenting brain blood vessels has become a crucial scientific challenge. We aimed to develop a fully automated deep learning workflow that achieves accurate 3D segmentation of cerebral blood vessels by incorporating classic convolutional neural networks (CNNs) and transformer models.
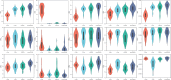
Methods: We used a public cerebrovascular segmentation dataset (CSD) containing 45 volumes of 1.5 T time-of-flight magnetic resonance angiography images. We collected data from another private middle cerebral artery (MCA) with lenticulostriate artery (LSA) segmentation dataset (MLD), which encompassed 3.0 T three-dimensional T1-weighted sequences of volumetric isotropic turbo spin echo acquisition MRI images of 107 patients aged 62 ± 11 years (42 females). The workflow includes data analysis, preprocessing, augmentation, model training with validation, and postprocessing techniques. Brain vessels were segmented using the U-Net, V-Net, UNETR, and SwinUNETR models. The model performances were evaluated using the dice similarity coefficient (DSC), average surface distance (ASD), precision (PRE), sensitivity (SEN), and specificity (SPE).
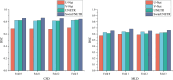
Results: During 4-fold cross-validation, SwinUNETR obtained the highest DSC in each fold. On the CSD test set, SwinUNETR achieved the best DSC (0.853), PRE (0.848), SEN (0.860), and SPE (0.9996), while V-Net achieved the best ASD (0.99). On the MLD test set, SwinUNETR demonstrated good MCA segmentation performance and had the best DSC, ASD, PRE, and SPE for segmenting the LSA.
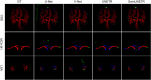
Conclusions: The workflow demonstrated excellent performance on different sequences of MRI images for vessels of varying sizes. This method allows doctors to visualize cerebrovascular structures.
Critical relevance statement: A deep learning-based 3D cerebrovascular segmentation workflow is feasible and promising for visualizing cerebrovascular structures and monitoring cerebral small vessels, such as lenticulostriate arteries.
Key points: • The proposed deep learning-based workflow performs well in cerebrovascular segmentation tasks. • Among comparison models, SwinUNETR achieved the best DSC, ASD, PRE, and SPE values in lenticulostriate artery segmentation. • The proposed workflow can be used for different MR sequences, such as bright and black blood imaging.
Keywords: Angiography; Black blood imaging; Cerebrovascular segmentation; Deep learning; Magnetic resonance.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Automatic segmentation and volume measurement of anterior visual pathway in brain 3D-T1WI using deep learning.Front Med (Lausanne). 2025 Apr 28;12:1530361. doi: 10.3389/fmed.2025.1530361. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40357297 Free PMC article.
-
Automated 3D U-net based segmentation of neonatal cerebral ventricles from 3D ultrasound images.Med Phys. 2022 Feb;49(2):1034-1046. doi: 10.1002/mp.15432. Epub 2022 Jan 12. Med Phys. 2022. PMID: 34958147
-
Two-stage deep learning model for fully automated pancreas segmentation on computed tomography: Comparison with intra-reader and inter-reader reliability at full and reduced radiation dose on an external dataset.Med Phys. 2021 May;48(5):2468-2481. doi: 10.1002/mp.14782. Epub 2021 Mar 16. Med Phys. 2021. PMID: 33595105
-
Vessel segmentation from volumetric images: a multi-scale double-pathway network with class-balanced loss at the voxel level.Med Phys. 2021 Jul;48(7):3804-3814. doi: 10.1002/mp.14934. Epub 2021 May 31. Med Phys. 2021. PMID: 33969487
-
Magnetic resonance image-based brain tumour segmentation methods: A systematic review.Digit Health. 2022 Mar 16;8:20552076221074122. doi: 10.1177/20552076221074122. eCollection 2022 Jan-Dec. Digit Health. 2022. PMID: 35340900 Free PMC article. Review.
References
-
- Sakata A, Fushimi Y, Okada T, et al. Evaluation of cerebral arteriovenous shunts: a comparison of parallel imaging time-of-flight magnetic resonance angiography (TOF-MRA) and compressed sensing TOF-MRA to digital subtraction angiography. Neuroradiology. 2021;63(6):879–887. doi: 10.1007/s00234-020-02581-y. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

