Evolution of chromosome-arm aberrations in breast cancer through genetic network rewiring
- PMID: 38517886
- PMCID: PMC11063629
- DOI: 10.1016/j.celrep.2024.113988
Evolution of chromosome-arm aberrations in breast cancer through genetic network rewiring
Abstract
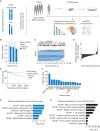
The basal breast cancer subtype is enriched for triple-negative breast cancer (TNBC) and displays consistent large chromosomal deletions. Here, we characterize evolution and maintenance of chromosome 4p (chr4p) loss in basal breast cancer. Analysis of The Cancer Genome Atlas data shows recurrent deletion of chr4p in basal breast cancer. Phylogenetic analysis of a panel of 23 primary tumor/patient-derived xenograft basal breast cancers reveals early evolution of chr4p deletion. Mechanistically we show that chr4p loss is associated with enhanced proliferation. Gene function studies identify an unknown gene, C4orf19, within chr4p, which suppresses proliferation when overexpressed-a member of the PDCD10-GCKIII kinase module we name PGCKA1. Genome-wide pooled overexpression screens using a barcoded library of human open reading frames identify chromosomal regions, including chr4p, that suppress proliferation when overexpressed in a context-dependent manner, implicating network interactions. Together, these results shed light on the early emergence of complex aneuploid karyotypes involving chr4p and adaptive landscapes shaping breast cancer genomes.
Keywords: CP: Cancer; CP: Genomics; GCK-III; PDCD10; aneuploidy; basal breast cancer; cancer evolution; chromosomal arm copy number aberrations; chromosome 4p; triple-negative breast cancer.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Comment in
-
Genomic linkages dictate cancer evolution.Cell Rep. 2024 May 28;43(5):114133. doi: 10.1016/j.celrep.2024.114133. Epub 2024 Apr 20. Cell Rep. 2024. PMID: 38643481
References
-
- Hammond M.E.H., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S., Fitzgibbons P.L., Francis G., Goldstein N.S., Hayes M., et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784–2795. - PMC - PubMed
-
- Haffty B.G., Yang Q., Reiss M., Kearney T., Higgins S.A., Weidhaas J., Harris L., Hait W., Toppmeyer D. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J. Clin. Oncol. 2006;24:5652–5657. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

