High-throughput sequencing of insect specimens with sub-optimal DNA preservation using a practical, plate-based Illumina-compatible Tn5 transposase library preparation method
- PMID: 38517905
- PMCID: PMC10959394
- DOI: 10.1371/journal.pone.0300865
High-throughput sequencing of insect specimens with sub-optimal DNA preservation using a practical, plate-based Illumina-compatible Tn5 transposase library preparation method
Abstract
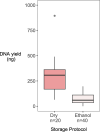
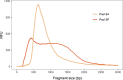
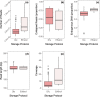
Entomological sampling and storage conditions often prioritise efficiency, practicality and conservation of morphological characteristics, and may therefore be suboptimal for DNA preservation. This practice can impact downstream molecular applications, such as the generation of high-throughput genomic libraries, which often requires substantial DNA input amounts. Here, we use a practical Tn5 transposase tagmentation-based library preparation method optimised for 96-well plates and low yield DNA extracts from insect legs that were stored under sub-optimal conditions for DNA preservation. The samples were kept in field vehicles for extended periods of time, before long-term storage in ethanol in the freezer, or dry at room temperature. By reducing DNA input to 6ng, more samples with sub-optimal DNA yields could be processed. We matched this low DNA input with a 6-fold dilution of a commercially available tagmentation enzyme, significantly reducing library preparation costs. Costs and workload were further suppressed by direct post-amplification pooling of individual libraries. We generated medium coverage (>3-fold) genomes for 88 out of 90 specimens, with an average of approximately 10-fold coverage. While samples stored in ethanol yielded significantly less DNA compared to those which were stored dry, these samples had superior sequencing statistics, with longer sequencing reads and higher rates of endogenous DNA. Furthermore, we find that the efficiency of tagmentation-based library preparation can be improved by a thorough post-amplification bead clean-up which selects against both short and large DNA fragments. By opening opportunities for the use of sub-optimally preserved, low yield DNA extracts, we broaden the scope of whole genome studies of insect specimens. We therefore expect these results and this protocol to be valuable for a range of applications in the field of entomology.
Copyright: © 2024 Cobb et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- van der Sluijs JP. Insect decline, an emerging global environmental risk. Current Opinion in Environmental Sustainability. 2020;46:39–42.
-
- Potts S, Dauber, J., Hochkirch, A., Oteman, B., Roy, D., Ahnre, K., et al. Proposal for an EU Pollinator Monitoring Scheme. Luxembourg; 2020. Report No.: ISBN 978-92-76-23859-1 Contract No.: JRC122225.
-
- Saunders ME, Janes JK, O’Hanlon JC. Moving On from the Insect Apocalypse Narrative: Engaging with Evidence-Based Insect Conservation. Bioscience. 2020;70(1):80–9.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

