Childhood cancer mutagenesis caused by transposase-derived PGBD5
- PMID: 38517960
- PMCID: PMC10959420
- DOI: 10.1126/sciadv.adn4649
Childhood cancer mutagenesis caused by transposase-derived PGBD5
Abstract
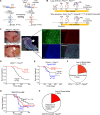
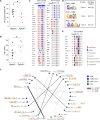
Genomic rearrangements are a hallmark of most childhood tumors, including medulloblastoma, one of the most common brain tumors in children, but their causes remain largely unknown. Here, we show that PiggyBac transposable element derived 5 (Pgbd5) promotes tumor development in multiple developmentally accurate mouse models of Sonic Hedgehog (SHH) medulloblastoma. Most Pgbd5-deficient mice do not develop tumors, while maintaining normal cerebellar development. Ectopic activation of SHH signaling is sufficient to enforce cerebellar granule cell progenitor-like cell states, which exhibit Pgbd5-dependent expression of distinct DNA repair and neurodevelopmental factors. Mouse medulloblastomas expressing Pgbd5 have increased numbers of somatic structural DNA rearrangements, some of which carry PGBD5-specific sequences at their breakpoints. Similar sequence breakpoints recurrently affect somatic DNA rearrangements of known tumor suppressors and oncogenes in medulloblastomas in 329 children. This identifies PGBD5 as a medulloblastoma mutator and provides a genetic mechanism for the generation of oncogenic DNA rearrangements in childhood cancer.
Figures



References
-
- Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). - PubMed
-
- Anderson N. D., de Borja R., Young M. D., Fuligni F., Rosic A., Roberts N. D., Hajjar S., Layeghifard M., Novokmet A., Kowalski P. E., Anaka M., Davidson S., Zarrei M., Id Said B., Schreiner L. C., Marchand R., Sitter J., Gokgoz N., Brunga L., Graham G. T., Fullam A., Pillay N., Toretsky J. A., Yoshida A., Shibata T., Metzler M., Somers G. R., Scherer S. W., Flanagan A. M., Campbell P. J., Schiffman J. D., Shago M., Alexandrov L. B., Wunder J. S., Andrulis I. L., Malkin D., Behjati S., Shlien A., Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 361, eaam8419 (2018). - PMC - PubMed
-
- Henssen A. G., Koche R., Zhuang J., Jiang E., Reed C., Eisenberg A., Still E., MacArthur I. C., Rodríguez-Fos E., Gonzalez S., Puiggròs M., Blackford A. N., Mason C. E., de Stanchina E., Gönen M., Emde A. K., Shah M., Arora K., Reeves C., Socci N. D., Perlman E., Antonescu C. R., Roberts C. W. M., Steen H., Mullen E., Jackson S. P., Torrents D., Weng Z., Armstrong S. A., Kentsis A., PGBD5 promotes site-specific oncogenic mutations in human tumors. Nat. Genet. 49, 1005–1014 (2017). - PMC - PubMed
-
- Henssen A. G., Reed C., Jiang E., Garcia H. D., von Stebut J., MacArthur I. C., Hundsdoerfer P., Kim J. H., de Stanchina E., Kuwahara Y., Hosoi H., Ganem N. J., dela Cruz F., Kung A. L., Schulte J. H., Petrini J. H., Kentsis A., Therapeutic targeting of PGBD5-induced DNA repair dependency in pediatric solid tumors. Sci. Transl. Med. 9, eaam9078 (2017). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

