Peripheral priming induces plastic transcriptomic and proteomic responses in circulating neutrophils required for pathogen containment
- PMID: 38517968
- PMCID: PMC11093074
- DOI: 10.1126/sciadv.adl1710
Peripheral priming induces plastic transcriptomic and proteomic responses in circulating neutrophils required for pathogen containment
Abstract
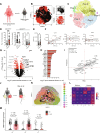
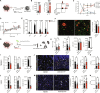
Neutrophils rapidly respond to inflammation and infection, but to which degree their functional trajectories after mobilization from the bone marrow are shaped within the circulation remains vague. Experimental limitations have so far hampered neutrophil research in human disease. Here, using innovative fixation and single-cell-based toolsets, we profile human and murine neutrophil transcriptomes and proteomes during steady state and bacterial infection. We find that peripheral priming of circulating neutrophils leads to dynamic shifts dominated by conserved up-regulation of antimicrobial genes across neutrophil substates, facilitating pathogen containment. We show the TLR4/NF-κB signaling-dependent up-regulation of canonical neutrophil activation markers like CD177/NB-1 during acute inflammation, resulting in functional shifts in vivo. Blocking de novo RNA synthesis in circulating neutrophils abrogates these plastic shifts and prevents the adaptation of antibacterial neutrophil programs by up-regulation of distinct effector molecules upon infection. These data underline transcriptional plasticity as a relevant mechanism of functional neutrophil reprogramming during acute infection to foster bacterial containment within the circulation.
Figures






References
-
- Ley K., Hoffman H. M., Kubes P., Cassatella M. A., Zychlinsky A., Hedrick C. C., Catz S. D., Neutrophils: New insights and open questions. Sci. Immunol. 3, eaat4579 (2018). - PubMed
-
- Kolaczkowska E., Kubes P., Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

