Arginine methylation of SM-LIKE PROTEIN 4 antagonistically affects alternative splicing during Arabidopsis stress responses
- PMID: 38518124
- PMCID: PMC11132874
- DOI: 10.1093/plcell/koae051
Arginine methylation of SM-LIKE PROTEIN 4 antagonistically affects alternative splicing during Arabidopsis stress responses
Abstract
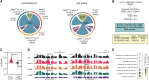
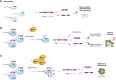
Arabidopsis (Arabidopsis thaliana) PROTEIN ARGININE METHYLTRANSFERASE5 (PRMT5) post-translationally modifies RNA-binding proteins by arginine (R) methylation. However, the impact of this modification on the regulation of RNA processing is largely unknown. We used the spliceosome component, SM-LIKE PROTEIN 4 (LSM4), as a paradigm to study the role of R-methylation in RNA processing. We found that LSM4 regulates alternative splicing (AS) of a suite of its in vivo targets identified here. The lsm4 and prmt5 mutants show a considerable overlap of genes with altered AS raising the possibility that splicing of those genes could be regulated by PRMT5-dependent LSM4 methylation. Indeed, LSM4 methylation impacts AS, particularly of genes linked with stress response. Wild-type LSM4 and an unmethylable version complement the lsm4-1 mutant, suggesting that methylation is not critical for growth in normal environments. However, LSM4 methylation increases with abscisic acid and is necessary for plants to grow under abiotic stress. Conversely, bacterial infection reduces LSM4 methylation, and plants that express unmethylable-LSM4 are more resistant to Pseudomonas than those expressing wild-type LSM4. This tolerance correlates with decreased intron retention of immune-response genes upon infection. Taken together, this provides direct evidence that R-methylation adjusts LSM4 function on pre-mRNA splicing in an antagonistic manner in response to biotic and abiotic stress.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

