Mitophagy mediated by BNIP3 and NIX protects against ferroptosis by downregulating mitochondrial reactive oxygen species
- PMID: 38519771
- PMCID: PMC11094013
- DOI: 10.1038/s41418-024-01280-y
Mitophagy mediated by BNIP3 and NIX protects against ferroptosis by downregulating mitochondrial reactive oxygen species
Abstract
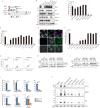
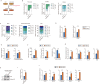
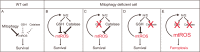
Mitophagy plays an important role in the maintenance of mitochondrial homeostasis and can be categorized into two types: ubiquitin-mediated and receptor-mediated pathways. During receptor-mediated mitophagy, mitophagy receptors facilitate mitophagy by tethering the isolation membrane to mitochondria. Although at least five outer mitochondrial membrane proteins have been identified as mitophagy receptors, their individual contribution and interrelationship remain unclear. Here, we show that HeLa cells lacking BNIP3 and NIX, two of the five receptors, exhibit a complete loss of mitophagy in various conditions. Conversely, cells deficient in the other three receptors show normal mitophagy. Using BNIP3/NIX double knockout (DKO) cells as a model, we reveal that mitophagy deficiency elevates mitochondrial reactive oxygen species (mtROS), which leads to activation of the Nrf2 antioxidant pathway. Notably, BNIP3/NIX DKO cells are highly sensitive to ferroptosis when Nrf2-driven antioxidant enzymes are compromised. Moreover, the sensitivity of BNIP3/NIX DKO cells is fully rescued upon the introduction of wild-type BNIP3 and NIX, but not the mutant forms incapable of facilitating mitophagy. Consequently, our results demonstrate that BNIP3 and NIX-mediated mitophagy plays a role in regulating mtROS levels and protects cells from ferroptosis.
© 2024. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
- 19H05712/MEXT | Japan Society for the Promotion of Science (JSPS)
- 18H04858/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP21gm6110013/Japan Agency for Medical Research and Development (AMED)
- 16KK0162/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22K07207/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

